 |
PDBsum entry 1p4r

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase, hydrolase
|
PDB id
|
|

|

|
1p4r
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.1.2.3
- phosphoribosylaminoimidazolecarboxamide formyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Purine Biosynthesis (late stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(6R)-10-formyltetrahydrofolate + 5-amino-1-(5-phospho-beta-D- ribosyl)imidazole-4-carboxamide = 5-formamido-1-(5-phospho-D- ribosyl)imidazole-4-carboxamide + (6S)-5,6,7,8-tetrahydrofolate
|
 |
 |
 |
 |
 |
(6R)-10-formyltetrahydrofolate
|
+
|
5-amino-1-(5-phospho-beta-D- ribosyl)imidazole-4-carboxamide
|
=
|
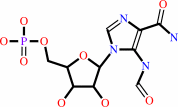 5-formamido-1-(5-phospho-D- ribosyl)imidazole-4-carboxamide
5-formamido-1-(5-phospho-D- ribosyl)imidazole-4-carboxamide
|
+
|
(6S)-5,6,7,8-tetrahydrofolate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.5.4.10
- Imp cyclohydrolase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + H2O = 5-formamido-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide
|
 |
 |
 |
 |
 |
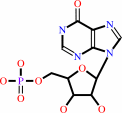 IMP
IMP
|
+
|
H2O
|
=
|
5-formamido-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:18034-18045
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of human bifunctional enzyme aminoimidazole-4-carboxamide ribonucleotide transformylase/IMP cyclohydrolase in complex with potent sulfonyl-containing antifolates.
|
|
C.G.Cheong,
D.W.Wolan,
S.E.Greasley,
P.A.Horton,
G.P.Beardsley,
I.A.Wilson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Aminoimidazole-4-carboxamide ribonucleotide (AICAR) transformylase/IMP
cyclohydrolase (ATIC) is a bifunctional enzyme with folate-dependent AICAR
transformylase and IMP cyclohydrolase activities that catalyzes the last two
steps of purine biosynthesis. The AICAR transformylase inhibitors BW1540 and
BW2315 are sulfamido-bridged 5,8-dideazafolate analogs with remarkably potent
K(i) values of 8 and 6 nm, respectively, compared with most other antifolates.
Crystal structures of ATIC at 2.55 and 2.60 A with each inhibitor, in the
presence of substrate AICAR, revealed that the sulfonyl groups dominate
inhibitor binding and orientation through interaction with the proposed oxyanion
hole. These agents then appear to mimic the anionic transition state and now
implicate Asn(431') in the reaction mechanism along with previously identified
key catalytic residues Lys(266) and His(267). Potent and selective inhibition of
the AICAR transformylase active site, compared with other folate-dependent
enzymes, should therefore be pursued by further design of sulfonyl-containing
antifolates.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
FIG. 4. Comparison of the interactions of the AICAR Tfase
active site with BW1540 and BW2315. A, schematic representation
of the hydrogen bonding network and corresponding distances
within the active site between AICAR, BW1540 and the AICAR Tfase
residues. Interacting residues from the opposite subunit of the
AICAR-bound monomer (black) are labeled in blue and indicated
with a prime symbol. B, schematic representation of the hydrogen
bonding network and corresponding distances within the AICAR
Tfase active site with the bound AICAR and BW2315 molecules.
Interacting residues from the opposite subunit of the
AICAR-bound monomer (black) are labeled in blue and are
indicated with a prime symbol.
|
 |
Figure 5.
FIG. 5. Comparison of free and bound AICAR Tfase active
sites. A, superposition of the AICAR- and antifolate-bound human
AICAR Tfase active sites reveals slight translational and
orientational deviations between the inhibitor molecules because
of the propensity for the sulfonyl groups to be located in the
oxyanion hole. The BW1540-bound structure is colored and labeled
according to Fig. 3A. BW2315 carbons are colored orange with the
BW2315-bound ATIC carbons and C  trace colored in wheat.
B, superposition of the BW1540-bound structure and the apo AICAR
Tfase active sites reveals the slight conformational changes
that occur upon folate binding. The BW1540-bound structure is
colored and labeled according to Fig. 3A with the apo human ATIC
C trace colored in wheat.
B, superposition of the BW1540-bound structure and the apo AICAR
Tfase active sites reveals the slight conformational changes
that occur upon folate binding. The BW1540-bound structure is
colored and labeled according to Fig. 3A with the apo human ATIC
C  trace colored in wheat. trace colored in wheat.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
18034-18045)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.D.Durrant,
R.E.Amaro,
and
J.A.McCammon
(2009).
AutoGrow: a novel algorithm for protein inhibitor design.
|
| |
Chem Biol Drug Des,
73,
168-178.
|
 |
|
|
|
|
 |
S.L.Cao,
Y.W.Guo,
X.B.Wang,
M.Zhang,
Y.P.Feng,
Y.Y.Jiang,
Y.Wang,
Q.Gao,
and
J.Ren
(2009).
Synthesis and cytotoxicity screening of piperazine-1-carbodithioate derivatives of 2-substituted quinazolin-4(3H)-ones.
|
| |
Arch Pharm (Weinheim),
342,
182-189.
|
 |
|
|
|
|
 |
Y.Zhang,
M.Morar,
and
S.E.Ealick
(2008).
Structural biology of the purine biosynthetic pathway.
|
| |
Cell Mol Life Sci,
65,
3699-3724.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

