 |
PDBsum entry 1ogc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.4.99.62
- D-ribose pyranase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-ribopyranose = beta-D-ribofuranose
|
 |
 |
 |
 |
 |
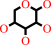 beta-D-ribopyranose
beta-D-ribopyranose
|
=
|
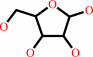 beta-D-ribofuranose
beta-D-ribofuranose
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
278:28173-28180
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of RbsD leading to the identification of cytoplasmic sugar-binding proteins with a novel folding architecture.
|
|
M.S.Kim,
J.Shin,
W.Lee,
H.S.Lee,
B.H.Oh.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
RbsD is the only protein whose biochemical function is unknown among the six
gene products of the rbs operon involved in the active transport of ribose.
FucU, a paralogue of RbsD conserved from bacteria to human, is also the only
protein whose function is unknown among the seven gene products of the l-fucose
regulon. Here we report the crystal structures of Bacillus subtilis RbsD, which
reveals a novel decameric toroidal assembly of the protein. Nuclear magnetic
resonance and other studies on RbsD reveal that the intersubunit cleft of the
protein binds specific forms of d-ribose, but it does not have an enzyme
activity toward the sugar. Likewise, FucU binds l-fucose but lacks an enzyme
activity toward this sugar. We conclude that RbsD and FucU are cytoplasmic
sugar-binding proteins, a novel class of proteins whose functional role may lie
in helping influx of the sugar substrates.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIG. 1. Structure and ribose-binding site of RbsD. a,
decameric assembly of RbsD shown in two different orientations.
Ribose molecules bound to the intersubunit clefts are shown as
ball-and-sticks. b, ribbon diagram of RbsD monomer. The
secondary structures are numbered in the order of appearance in
the primary sequence. c, buried ion cage. A negatively charged
ion (in cyan), which is putatively a Cl-, is bound between two
RbsD subunits related by the molecular 2-fold axis that
superimposes the pentameric rings. The symmetry-related pairs
are shown in different colors. Water molecules are in red. Two
histidine residues hydrogen-bonded to the bound water molecules
are shown.
|
 |
Figure 3.
FIG. 3. Binding of ribose and ribose 5-phosphate to RbsD.
a, binding of D-ribose. The  -pyranoside form of the
sugar is fitted into the final 2F[o] - F[c] map (1.95 Å,
1.1 -pyranoside form of the
sugar is fitted into the final 2F[o] - F[c] map (1.95 Å,
1.1  ). The C-4-OH group
interacts with the C-terminal carboxyl group via a water
molecule. The density of this water molecule is weak or
unobserved in 2 of 5 binding sites. These weak interactions are
not shown. b, surface representation of the intersubunit cleft.
The bound ribose is displayed in CPK mode with the oxygen and
carbon atoms in red and black, respectively. The bound water
molecule is in blue. c, binding of D-ribose 5-phosphate. The
phosphorylated sugar is in the ). The C-4-OH group
interacts with the C-terminal carboxyl group via a water
molecule. The density of this water molecule is weak or
unobserved in 2 of 5 binding sites. These weak interactions are
not shown. b, surface representation of the intersubunit cleft.
The bound ribose is displayed in CPK mode with the oxygen and
carbon atoms in red and black, respectively. The bound water
molecule is in blue. c, binding of D-ribose 5-phosphate. The
phosphorylated sugar is in the  -furanose form, which
was determined at the beginning of the refinement. The final
2F[o] - F[c] map (2.05 Å, 1.1 -furanose form, which
was determined at the beginning of the refinement. The final
2F[o] - F[c] map (2.05 Å, 1.1  ) is shown. His-20 from
one RbsD subunit is shown in cyan, and rest of the residues from
the adjacent subunit are shown in green. Figs. 1 and 3 were
prepared with the program Bobscript (26). ) is shown. His-20 from
one RbsD subunit is shown in cyan, and rest of the residues from
the adjacent subunit are shown in green. Figs. 1 and 3 were
prepared with the program Bobscript (26).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2003,
278,
28173-28180)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Wang,
M.Wu,
and
J.Zang
(2011).
Crystal structure of Sa240: A ribose pyranase homolog with partial active site from Staphylococcus aureus.
|
| |
J Struct Biol,
174,
413-419.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Feng,
M.Zhang,
M.Hu,
J.Zheng,
W.Jiao,
and
Z.Chang
(2009).
Disassembly intermediates of RbsD protein remain oligomeric despite the loss of an intact secondary structure.
|
| |
Sci China C Life Sci,
52,
997.
|
 |
|
|
|
|
 |
A.D.Hill,
and
P.J.Reilly
(2008).
A Gibbs free energy correlation for automated docking of carbohydrates.
|
| |
J Comput Chem,
29,
1131-1141.
|
 |
|
|
|
|
 |
J.S.Richardson,
X.Carpena,
J.Switala,
R.Perez-Luque,
L.J.Donald,
P.C.Loewen,
and
I.J.Oresnik
(2008).
RhaU of Rhizobium leguminosarum is a rhamnose mutarotase.
|
| |
J Bacteriol,
190,
2903-2910.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Zhou,
J.Boekhorst,
C.Francke,
and
R.J.Siezen
(2008).
LocateP: genome-scale subcellular-location predictor for bacterial proteins.
|
| |
BMC Bioinformatics,
9,
173.
|
 |
|
|
|
|
 |
Y.Feng,
W.Jiao,
X.Fu,
and
Z.Chang
(2006).
Stepwise disassembly and apparent nonstepwise reassembly for the oligomeric RbsD protein.
|
| |
Protein Sci,
15,
1441-1448.
|
 |
|
|
|
|
 |
L.Tomasinsig,
M.Scocchi,
R.Mettulio,
and
M.Zanetti
(2004).
Genome-wide transcriptional profiling of the Escherichia coli response to a proline-rich antimicrobial peptide.
|
| |
Antimicrob Agents Chemother,
48,
3260-3267.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

