 |
PDBsum entry 1of1

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.21
- thymidine kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
thymidine + ATP = dTMP + ADP + H+
|
 |
 |
 |
 |
 |
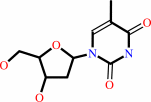 thymidine
thymidine
|
+
|
ATP
Bound ligand (Het Group name = )
matches with 84.21% similarity
|
=
|
 dTMP
dTMP
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:32832-32838
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Biochemical and structural characterization of (South)-methanocarbathymidine that specifically inhibits growth of herpes simplex virus type 1 thymidine kinase-transduced osteosarcoma cells.
|
|
P.Schelling,
M.T.Claus,
R.Johner,
V.E.Marquez,
G.E.Schulz,
L.Scapozza.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Two analogs of the natural nucleoside dT featuring a pseudosugar with fixed
conformation in place of the deoxyribosyl residue (carbathymidine analogs) were
biochemically and structurally characterized for their acceptance by both human
cytosolic thymidine kinase isoenzyme 1 (hTK1) and herpes simplex virus type 1
thymidine kinase (HSV1 TK) and subsequently tested in cell proliferation assays.
3'-exo-Methanocarbathymidine ((South)-methanocarbathymidine (S)-MCT), which is a
substrate for HSV1 TK, specifically inhibited growth of HSV1 TK-transduced human
osteosarcoma cells with an IC(50) value in the range of 15 microM without
significant toxicity toward both hTK1-negative (TK(-)) and non-transduced cells.
2'-exo-Methanocarbathymidine ((North)-methanocarbathymidine (N)-MCT), which is a
weak substrate for hTK1 and a substantial one for HSV1 TK, induced a specific
growth inhibition in HSV1 TK-transfected cells comparable to that of (S)-MCT and
ganciclovir. A growth inhibition activity was also observed with (N)-MCT and
ganciclovir in non-transduced cells in a cell line-dependent manner, whereas
TK(-) cells were not affected. The presented 1.95-A crystal structure of the
complex (S)-MCT.HSV1 TK explains both the more favorable binding affinity and
catalytic turnover of (S)-MCT for HSV1 TK over the North analog. Additionally
the plasticity of the active site of the enzyme is addressed by comparing the
binding of (North)- and (South)-carbathymidine analogs. The presented study of
these two potent candidate prodrugs for HSV1 TK gene-directed enzyme prodrug
therapy suggests that (S)-MCT may be even safer to use than its North
counterpart (N)-MCT.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIG. 1. Structures of (N)-MCT (A) and (S)-MCT (B).
|
 |
Figure 5.
FIG. 5. Stereoview of HSV1 TK structure in complex with
(S)-MCT. The conformation of the compound and the position of
the sulfate group are well defined by the electron density
contoured at 1.3  . The hydrogen bonding
patterns for the thymine moiety as well as for both 3'-OH and
5'-OH groups are the same as in the HSV1 TK·dT structure.
(S)-MCT and HSV1 TK carbon atoms are represented in orange and
black; nitrogen, oxygen, and sulfur atoms are in blue, red, and
yellow, respectively; and water molecules are represented as
green balls. H-bonds are depicted by dashed lines between donor
(D) and acceptor (A) and defined as follows: distance D--A,
2.8-3.2 Å; angle D-H--A, 140-180°. . The hydrogen bonding
patterns for the thymine moiety as well as for both 3'-OH and
5'-OH groups are the same as in the HSV1 TK·dT structure.
(S)-MCT and HSV1 TK carbon atoms are represented in orange and
black; nitrogen, oxygen, and sulfur atoms are in blue, red, and
yellow, respectively; and water molecules are represented as
green balls. H-bonds are depicted by dashed lines between donor
(D) and acceptor (A) and defined as follows: distance D--A,
2.8-3.2 Å; angle D-H--A, 140-180°.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
32832-32838)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.Y.Michel,
and
P.Strazewski
(2009).
Total syntheses of a conformationally locked North-type methanocarba puromycin analogue and a dinucleotide derivative.
|
| |
Chemistry,
15,
6244-6257.
|
 |
|
|
|
|
 |
Y.Mehellou,
J.Balzarini,
and
C.McGuigan
(2009).
An investigation into the anti-HIV activity of 2',3'-didehydro-2',3'-dideoxyuridine (d4U) and 2',3'-dideoxyuridine (ddU) phosphoramidate 'ProTide' derivatives.
|
| |
Org Biomol Chem,
7,
2548-2553.
|
 |
|
|
|
|
 |
A.Melman,
M.Zhong,
V.E.Marquez,
and
K.A.Jacobson
(2008).
Synthesis of enantiomerically pure (S)-methanocarbaribo uracil nucleoside derivatives for use as antiviral agents and P2Y receptor ligands.
|
| |
J Org Chem,
73,
8085-8088.
|
 |
|
|
|
|
 |
D.F.Smee,
B.L.Hurst,
M.H.Wong,
R.I.Glazer,
A.Rahman,
and
R.W.Sidwell
(2007).
Efficacy of N-methanocarbathymidine in treating mice infected intranasally with the IHD and WR strains of vaccinia virus.
|
| |
Antiviral Res,
76,
124-129.
|
 |
|
|
|
|
 |
A.Johayem,
S.Raić-Malić,
K.Lazzati,
P.A.Schubiger,
L.Scapozza,
and
S.M.Ametamey
(2006).
Synthesis and characterization of a C6 nucleoside analogue for the in vivo imaging of the gene expression of herpes simplex virus type-1 thymidine kinase (HSV1 TK).
|
| |
Chem Biodivers,
3,
274-283.
|
 |
|
|
|
|
 |
K.El Omari,
N.Solaroli,
A.Karlsson,
J.Balzarini,
and
D.K.Stammers
(2006).
Structure of vaccinia virus thymidine kinase in complex with dTTP: insights for drug design.
|
| |
BMC Struct Biol,
6,
22.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.N.Prichard,
K.A.Keith,
D.C.Quenelle,
and
E.R.Kern
(2006).
Activity and mechanism of action of N-methanocarbathymidine against herpesvirus and orthopoxvirus infections.
|
| |
Antimicrob Agents Chemother,
50,
1336-1341.
|
 |
|
|
|
|
 |
W.Zhu,
A.Burnette,
D.Dorjsuren,
P.E.Roberts,
M.Huleihel,
R.H.Shoemaker,
V.E.Marquez,
R.Agbaria,
and
S.Sei
(2005).
Potent antiviral activity of north-methanocarbathymidine against Kaposi's sarcoma-associated herpesvirus.
|
| |
Antimicrob Agents Chemother,
49,
4965-4973.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

