 |
PDBsum entry 1miv

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Translation, transferase
|
PDB id
|
|

|

|
1miv
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.7.72
- Cca tRNA nucleotidyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
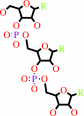
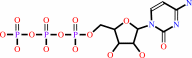
a tRNA precursor + 2 CTP + ATP = a tRNA with a 3' CCA end + 3 diphosphate
|
 |
 |
 |
 |
 |
 tRNA precursor
tRNA precursor
|
+
|
 2
×
CTP
2
×
CTP
|
+
|
 ATP
ATP
|
=
|
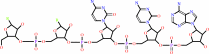 tRNA with a 3' CCA end
tRNA with a 3' CCA end
|
+
|
 3
×
diphosphate
3
×
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Cell
111:815-824
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP.
|
|
F.Li,
Y.Xiong,
J.Wang,
H.D.Cho,
K.Tomita,
A.M.Weiner,
T.A.Steitz.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
CCA-adding enzymes polymerize CCA onto the 3' terminus of immature tRNAs without
using a nucleic acid template. The 3.0 A resolution crystal structures of the
CCA-adding enzyme from Bacillus stearothermophilus and its complexes with ATP or
CTP reveal a seahorse-shaped subunit consisting of four domains: head, neck,
body, and tail. The head is structurally homologous to the palm domain of DNA
polymerase beta but has additional structural features and functions. The neck,
body, and tail represent new protein folding motifs. The neck provides a
specific template for the incoming ATP or CTP, whereas the body and tail may
bind tRNA. Each subunit has one active site capable of switching its base
specificity between ATP and CTP, an important component of the CCA-adding
mechanism.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Important Interactions between ATP and Conserved
Residues of BstCCA that Confer Specificity for the Base, the
Ribose, and the TriphosphateThe five conserved sequence motifs,
as explained in detail in Figure 5, are: A (magenta), B (light
green), C (dark green), D (yellow), and E (brown).
|
 |
Figure 4.
Figure 4. Base-Specific Interactions between BstCCA and ATP
or CTP(A) Interactions between the carboxylate of D154 and the
guanidinium group of R157 of BstCCA and ATP.(B) Interaction
between D154 and R157 of BstCCA and CTP. The position of the
guanidinium group of R157 is changed to be complementary to C
while D154 interacts with the N4 of C in much the same way as it
interacts with the N6 of A.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Cell
(2002,
111,
815-824)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Hoffmeier,
H.Betat,
A.Bluschke,
R.Günther,
S.Junghanns,
H.J.Hofmann,
and
M.Mörl
(2010).
Unusual evolution of a catalytic core element in CCA-adding enzymes.
|
| |
Nucleic Acids Res,
38,
4436-4447.
|
 |
|
|
|
|
 |
B.Pan,
Y.Xiong,
and
T.A.Steitz
(2010).
How the CCA-adding enzyme selects adenine over cytosine at position 76 of tRNA.
|
| |
Science,
330,
937-940.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Betat,
C.Rammelt,
and
M.Mörl
(2010).
tRNA nucleotidyltransferases: ancient catalysts with an unusual mechanism of polymerization.
|
| |
Cell Mol Life Sci,
67,
1447-1463.
|
 |
|
|
|
|
 |
I.Y.Morozov,
M.G.Jones,
A.A.Razak,
D.J.Rigden,
and
M.X.Caddick
(2010).
CUCU modification of mRNA promotes decapping and transcript degradation in Aspergillus nidulans.
|
| |
Mol Cell Biol,
30,
460-469.
|
 |
|
|
|
|
 |
J.Yamtich,
and
J.B.Sweasy
(2010).
DNA polymerase family X: function, structure, and cellular roles.
|
| |
Biochim Biophys Acta,
1804,
1136-1150.
|
 |
|
|
|
|
 |
M.G.Abad,
B.S.Rao,
and
J.E.Jackman
(2010).
Template-dependent 3'-5' nucleotide addition is a shared feature of tRNAHis guanylyltransferase enzymes from multiple domains of life.
|
| |
Proc Natl Acad Sci U S A,
107,
674-679.
|
 |
|
|
|
|
 |
Y.M.Hou
(2010).
CCA addition to tRNA: implications for tRNA quality control.
|
| |
IUBMB Life,
62,
251-260.
|
 |
|
|
|
|
 |
S.Kim,
C.Liu,
K.Halkidis,
H.B.Gamper,
and
Y.M.Hou
(2009).
Distinct kinetic determinants for the stepwise CCA addition to tRNA.
|
| |
RNA,
15,
1827-1836.
|
 |
|
|
|
|
 |
Y.Toh,
D.Takeshita,
T.Numata,
S.Fukai,
O.Nureki,
and
K.Tomita
(2009).
Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme.
|
| |
EMBO J,
28,
3353-3365.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Just,
F.Butter,
M.Trenkmann,
T.Heitkam,
M.Mörl,
and
H.Betat
(2008).
A comparative analysis of two conserved motifs in bacterial poly(A) polymerase and CCA-adding enzyme.
|
| |
Nucleic Acids Res,
36,
5212-5220.
|
 |
|
|
|
|
 |
A.Neuenfeldt,
A.Just,
H.Betat,
and
M.Mörl
(2008).
Evolution of tRNA nucleotidyltransferases: a small deletion generated CC-adding enzymes.
|
| |
Proc Natl Acad Sci U S A,
105,
7953-7958.
|
 |
|
|
|
|
 |
G.Martin,
S.Doublié,
and
W.Keller
(2008).
Determinants of substrate specificity in RNA-dependent nucleotidyl transferases.
|
| |
Biochim Biophys Acta,
1779,
206-216.
|
 |
|
|
|
|
 |
M.Dupasquier,
S.Kim,
K.Halkidis,
H.Gamper,
and
Y.M.Hou
(2008).
tRNA integrity is a prerequisite for rapid CCA addition: implication for quality control.
|
| |
J Mol Biol,
379,
579-588.
|
 |
|
|
|
|
 |
X.Shan,
T.A.Russell,
S.M.Paul,
D.B.Kushner,
and
P.B.Joyce
(2008).
Characterization of a temperature-sensitive mutation that impairs the function of yeast tRNA nucleotidyltransferase.
|
| |
Yeast,
25,
219-233.
|
 |
|
|
|
|
 |
Y.Toh,
T.Numata,
K.Watanabe,
D.Takeshita,
O.Nureki,
and
K.Tomita
(2008).
Molecular basis for maintenance of fidelity during the CCA-adding reaction by a CCA-adding enzyme.
|
| |
EMBO J,
27,
1944-1952.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Martin,
and
W.Keller
(2007).
RNA-specific ribonucleotidyl transferases.
|
| |
RNA,
13,
1834-1849.
|
 |
|
|
|
|
 |
H.D.Cho,
C.L.Verlinde,
and
A.M.Weiner
(2007).
Reengineering CCA-adding enzymes to function as (U,G)- or dCdCdA-adding enzymes or poly(C,A) and poly(U,G) polymerases.
|
| |
Proc Natl Acad Sci U S A,
104,
54-59.
|
 |
|
|
|
|
 |
J.Stagno,
I.Aphasizheva,
A.Rosengarth,
H.Luecke,
and
R.Aphasizhev
(2007).
UTP-bound and Apo structures of a minimal RNA uridylyltransferase.
|
| |
J Mol Biol,
366,
882-899.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.D.Cho,
Y.Chen,
G.Varani,
and
A.M.Weiner
(2006).
A model for C74 addition by CCA-adding enzymes: C74 addition, like C75 and A76 addition, does not involve tRNA translocation.
|
| |
J Biol Chem,
281,
9801-9811.
|
 |
|
|
|
|
 |
H.Oshikane,
K.Sheppard,
S.Fukai,
Y.Nakamura,
R.Ishitani,
T.Numata,
R.L.Sherrer,
L.Feng,
E.Schmitt,
M.Panvert,
S.Blanquet,
Y.Mechulam,
D.Söll,
and
O.Nureki
(2006).
Structural basis of RNA-dependent recruitment of glutamine to the genetic code.
|
| |
Science,
312,
1950-1954.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Suzuki,
P.Babitzke,
S.R.Kushner,
and
T.Romeo
(2006).
Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E.
|
| |
Genes Dev,
20,
2605-2617.
|
 |
|
|
|
|
 |
L.Wang,
J.Xie,
and
P.G.Schultz
(2006).
Expanding the genetic code.
|
| |
Annu Rev Biophys Biomol Struct,
35,
225-249.
|
 |
|
|
|
|
 |
B.H.Kim,
R.Sadreyev,
and
N.V.Grishin
(2005).
COG4849 is a novel family of nucleotidyltransferases.
|
| |
J Mol Recognit,
18,
422-425.
|
 |
|
|
|
|
 |
H.D.Cho,
C.L.Verlinde,
and
A.M.Weiner
(2005).
Archaeal CCA-adding enzymes: central role of a highly conserved beta-turn motif in RNA polymerization without translocation.
|
| |
J Biol Chem,
280,
9555-9566.
|
 |
|
|
|
|
 |
J.Deng,
N.L.Ernst,
S.Turley,
K.D.Stuart,
and
W.G.Hol
(2005).
Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei.
|
| |
EMBO J,
24,
4007-4017.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Bralley,
S.A.Chang,
and
G.H.Jones
(2005).
A phylogeny of bacterial RNA nucleotidyltransferases: Bacillus halodurans contains two tRNA nucleotidyltransferases.
|
| |
J Bacteriol,
187,
5927-5936.
|
 |
|
|
|
|
 |
A.F.Yakunin,
M.Proudfoot,
E.Kuznetsova,
A.Savchenko,
G.Brown,
C.H.Arrowsmith,
and
A.M.Edwards
(2004).
The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2',3'-cyclic phosphodiesterase, 2'-nucleotidase, and phosphatase activities.
|
| |
J Biol Chem,
279,
36819-36827.
|
 |
|
|
|
|
 |
A.M.Weiner
(2004).
tRNA maturation: RNA polymerization without a nucleic acid template.
|
| |
Curr Biol,
14,
R883-R885.
|
 |
|
|
|
|
 |
G.Martin,
and
W.Keller
(2004).
Sequence motifs that distinguish ATP(CTP):tRNA nucleotidyl transferases from eubacterial poly(A) polymerases.
|
| |
RNA,
10,
899-906.
|
 |
|
|
|
|
 |
H.D.Cho,
and
A.M.Weiner
(2004).
A single catalytically active subunit in the multimeric Sulfolobus shibatae CCA-adding enzyme can carry out all three steps of CCA addition.
|
| |
J Biol Chem,
279,
40130-40136.
|
 |
|
|
|
|
 |
I.Aphasizheva,
R.Aphasizhev,
and
L.Simpson
(2004).
RNA-editing terminal uridylyl transferase 1: identification of functional domains by mutational analysis.
|
| |
J Biol Chem,
279,
24123-24130.
|
 |
|
|
|
|
 |
K.Tomita,
S.Fukai,
R.Ishitani,
T.Ueda,
N.Takeuchi,
D.G.Vassylyev,
and
O.Nureki
(2004).
Structural basis for template-independent RNA polymerization.
|
| |
Nature,
430,
700-704.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Levinger,
M.Mörl,
and
C.Florentz
(2004).
Mitochondrial tRNA 3' end metabolism and human disease.
|
| |
Nucleic Acids Res,
32,
5430-5441.
|
 |
|
|
|
|
 |
L.Wang,
and
P.G.Schultz
(2004).
Expanding the genetic code.
|
| |
Angew Chem Int Ed Engl,
44,
34-66.
|
 |
|
|
|
|
 |
P.Schimmel,
and
X.L.Yang
(2004).
Two classes give lessons about CCA.
|
| |
Nat Struct Mol Biol,
11,
807-808.
|
 |
|
|
|
|
 |
T.Fujimura,
and
R.Esteban
(2004).
Bipartite 3'-cis-acting signal for replication in yeast 23 S RNA virus and its repair.
|
| |
J Biol Chem,
279,
13215-13223.
|
 |
|
|
|
|
 |
Y.Xiong,
and
T.A.Steitz
(2004).
Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template.
|
| |
Nature,
430,
640-645.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.D.Cho,
A.K.Oyelere,
S.A.Strobel,
and
A.M.Weiner
(2003).
Use of nucleotide analogs by class I and class II CCA-adding enzymes (tRNA nucleotidyltransferase): deciphering the basis for nucleotide selection.
|
| |
RNA,
9,
970-981.
|
 |
|
|
|
|
 |
M.Okabe,
K.Tomita,
R.Ishitani,
R.Ishii,
N.Takeuchi,
F.Arisaka,
O.Nureki,
and
S.Yokoyama
(2003).
Divergent evolutions of trinucleotide polymerization revealed by an archaeal CCA-adding enzyme structure.
|
| |
EMBO J,
22,
5918-5927.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Rott,
G.Zipor,
V.Portnoy,
V.Liveanu,
and
G.Schuster
(2003).
RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli.
|
| |
J Biol Chem,
278,
15771-15777.
|
 |
|
|
|
|
 |
Y.Xiong,
F.Li,
J.Wang,
A.M.Weiner,
and
T.A.Steitz
(2003).
Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes.
|
| |
Mol Cell,
12,
1165-1172.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

