 |
PDBsum entry 1m7v

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1m7v
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.14.47
- nitric-oxide synthase (flavodoxin).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3 reduced [flavodoxin] + 2 L-arginine + 4 O2 = 3 oxidized [flavodoxin] + 2 L-citrulline + 2 nitric oxide + 4 H2O + 5 H+
|
 |
 |
 |
 |
 |
3
×
reduced [flavodoxin]
|
+
|
2
×
L-arginine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 4
×
O2
4
×
O2
|
=
|
3
×
oxidized [flavodoxin]
|
+
|
 2
×
L-citrulline
2
×
L-citrulline
|
+
|
 2
×
nitric oxide
2
×
nitric oxide
|
+
|
4
×
H2O
|
+
|
5
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
5,6,7,8-tetrahydrobiopterin; Ferriheme b
|
 |
 |
 |
 |
 |
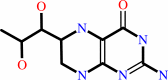 5,6,7,8-tetrahydrobiopterin
5,6,7,8-tetrahydrobiopterin
|
Ferriheme b
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
41:11071-11079
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of a nitric oxide synthase heme protein from Bacillus subtilis.
|
|
K.Pant,
A.M.Bilwes,
S.Adak,
D.J.Stuehr,
B.R.Crane.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Eukaryotic nitric oxide synthases (NOSs) produce nitric oxide to mediate
intercellular signaling and protect against pathogens. Recently, proteins
homologous to mammalian NOS oxygenase domains have been found in prokaryotes and
one from Bacillus subtilis (bsNOS) has been demonstrated to produce nitric oxide
[Adak, S., Aulak, K. S., and Stuehr, D. J. (2002) J. Biol. Chem. 277,
16167-16171]. We present structures of bsNOS complexed with the active cofactor
tetrahydrofolate and the substrate L-arginine (L-Arg) or the intermediate
N(omega)-hydroxy-L-arginine (NHA) to 1.9 or 2.2 A resolution, respectively. The
bsNOS structure is similar to those of the mammalian NOS oxygenase domains
(mNOS(ox)) except for the absence of an N-terminal beta-hairpin hook and
zinc-binding region that interact with pterin and stabilize the mNOS(ox) dimer.
Changes in patterns of residue conservation between bacterial and mammalian NOSs
correlate to different binding modes for pterin side chains. Residue
conservation on a surface patch surrounding an exposed heme edge indicates a
likely interaction site for reductase proteins in all NOSs. The heme pockets of
bsNOS and mNOS(ox) recognize L-Arg and NHA similarly, although a change from Val
to Ile beside the substrate guanidinium may explain the 10-20-fold slower
dissociation of product NO from the bacterial enzyme. Overall, these structures
suggest that bsNOS functions naturally to produce nitrogen oxides from L-Arg and
NHA in a pterin-dependent manner, but that the regulation and purpose of NO
production by NOS may be quite different in B. subtilis than in mammals.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Maréchal,
T.A.Mattioli,
D.J.Stuehr,
and
J.Santolini
(2010).
NO synthase isoforms specifically modify peroxynitrite reactivity.
|
| |
FEBS J,
277,
3963-3973.
|
 |
|
|
|
|
 |
B.R.Crane,
J.Sudhamsu,
and
B.A.Patel
(2010).
Bacterial nitric oxide synthases.
|
| |
Annu Rev Biochem,
79,
445-470.
|
 |
|
|
|
|
 |
S.R.Vincent
(2010).
Nitric oxide neurons and neurotransmission.
|
| |
Prog Neurobiol,
90,
246-255.
|
 |
|
|
|
|
 |
B.A.Patel,
M.Moreau,
J.Widom,
H.Chen,
L.Yin,
Y.Hua,
and
B.R.Crane
(2009).
Endogenous nitric oxide regulates the recovery of the radiation-resistant bacterium Deinococcus radiodurans from exposure to UV light.
|
| |
Proc Natl Acad Sci U S A,
106,
18183-18188.
|
 |
|
|
|
|
 |
I.Gusarov,
K.Shatalin,
M.Starodubtseva,
and
E.Nudler
(2009).
Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics.
|
| |
Science,
325,
1380-1384.
|
 |
|
|
|
|
 |
J.Sudhamsu,
and
B.R.Crane
(2009).
Bacterial nitric oxide synthases: what are they good for?
|
| |
Trends Microbiol,
17,
212-218.
|
 |
|
|
|
|
 |
T.Agapie,
S.Suseno,
J.J.Woodward,
S.Stoll,
R.D.Britt,
and
M.A.Marletta
(2009).
NO formation by a catalytically self-sufficient bacterial nitric oxide synthase from Sorangium cellulosum.
|
| |
Proc Natl Acad Sci U S A,
106,
16221-16226.
|
 |
|
|
|
|
 |
I.Gusarov,
M.Starodubtseva,
Z.Q.Wang,
L.McQuade,
S.J.Lippard,
D.J.Stuehr,
and
E.Nudler
(2008).
Bacterial nitric-oxide synthases operate without a dedicated redox partner.
|
| |
J Biol Chem,
283,
13140-13147.
|
 |
|
|
|
|
 |
K.Shatalin,
I.Gusarov,
E.Avetissova,
Y.Shatalina,
L.E.McQuade,
S.J.Lippard,
and
E.Nudler
(2008).
Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages.
|
| |
Proc Natl Acad Sci U S A,
105,
1009-1013.
|
 |
|
|
|
|
 |
N.J.Gilberthorpe,
and
R.K.Poole
(2008).
Nitric oxide homeostasis in Salmonella typhimurium: roles of respiratory nitrate reductase and flavohemoglobin.
|
| |
J Biol Chem,
283,
11146-11154.
|
 |
|
|
|
|
 |
A.W.Munro,
H.M.Girvan,
and
K.J.McLean
(2007).
Variations on a (t)heme--novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily.
|
| |
Nat Prod Rep,
24,
585-609.
|
 |
|
|
|
|
 |
F.J.Chartier,
and
M.Couture
(2007).
Substrate-specific interactions with the heme-bound oxygen molecule of nitric-oxide synthase.
|
| |
J Biol Chem,
282,
20877-20886.
|
 |
|
|
|
|
 |
H.K.Leiros,
A.L.Pey,
M.Innselset,
E.Moe,
I.Leiros,
I.H.Steen,
and
A.Martinez
(2007).
Structure of phenylalanine hydroxylase from Colwellia psychrerythraea 34H, a monomeric cold active enzyme with local flexibility around the active site and high overall stability.
|
| |
J Biol Chem,
282,
21973-21986.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.Q.Wang,
R.J.Lawson,
M.R.Buddha,
C.C.Wei,
B.R.Crane,
A.W.Munro,
and
D.J.Stuehr
(2007).
Bacterial flavodoxins support nitric oxide production by Bacillus subtilis nitric-oxide synthase.
|
| |
J Biol Chem,
282,
2196-2202.
|
 |
|
|
|
|
 |
F.J.Chartier,
S.P.Blais,
and
M.Couture
(2006).
A weak Fe-O bond in the oxygenated complex of the nitric-oxide synthase of Staphylococcus aureus.
|
| |
J Biol Chem,
281,
9953-9962.
|
 |
|
|
|
|
 |
J.Sudhamsu,
and
B.R.Crane
(2006).
Structure and reactivity of a thermostable prokaryotic nitric-oxide synthase that forms a long-lived oxy-heme complex.
|
| |
J Biol Chem,
281,
9623-9632.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Loria,
J.Kers,
and
M.Joshi
(2006).
Evolution of plant pathogenicity in Streptomyces.
|
| |
Annu Rev Phytopathol,
44,
469-487.
|
 |
|
|
|
|
 |
R.Pejchal,
E.Campbell,
B.D.Guenther,
B.W.Lennon,
R.G.Matthews,
and
M.L.Ludwig
(2006).
Structural perturbations in the Ala --> Val polymorphism of methylenetetrahydrofolate reductase: how binding of folates may protect against inactivation.
|
| |
Biochemistry,
45,
4808-4818.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.J.Chartier,
and
M.Couture
(2004).
Stability of the heme environment of the nitric oxide synthase from Staphylococcus aureus in the absence of pterin cofactor.
|
| |
Biophys J,
87,
1939-1950.
|
 |
|
|
|
|
 |
M.R.Buddha,
K.M.Keery,
and
B.R.Crane
(2004).
An unusual tryptophanyl tRNA synthetase interacts with nitric oxide synthase in Deinococcus radiodurans.
|
| |
Proc Natl Acad Sci U S A,
101,
15881-15886.
|
 |
|
|
|
|
 |
M.R.Buddha,
T.Tao,
R.J.Parry,
and
B.R.Crane
(2004).
Regioselective nitration of tryptophan by a complex between bacterial nitric-oxide synthase and tryptophanyl-tRNA synthetase.
|
| |
J Biol Chem,
279,
49567-49570.
|
 |
|
|
|
|
 |
Y.Sasaki,
N.Takaya,
A.Nakamura,
and
H.Shoun
(2004).
Isolation of flavohemoglobin from the actinomycete Streptomyces antibioticus grown without external nitric oxide stress.
|
| |
Biosci Biotechnol Biochem,
68,
1106-1112.
|
 |
|
|
|
|
 |
Z.Q.Wang,
C.C.Wei,
M.Sharma,
K.Pant,
B.R.Crane,
and
D.J.Stuehr
(2004).
A conserved Val to Ile switch near the heme pocket of animal and bacterial nitric-oxide synthases helps determine their distinct catalytic profiles.
|
| |
J Biol Chem,
279,
19018-19025.
|
 |
|
|
|
|
 |
K.Panda,
S.Adak,
K.S.Aulak,
J.Santolini,
J.F.McDonald,
and
D.J.Stuehr
(2003).
Distinct influence of N-terminal elements on neuronal nitric-oxide synthase structure and catalysis.
|
| |
J Biol Chem,
278,
37122-37131.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

