 |
PDBsum entry 1lkd

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1lkd
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of 2,3-dihydroxybiphenyl 1,2-dioxygenase (dhbd) complexed with 2',6'-dicl dihydroxybiphenyl (dhb)
|
|
Structure:
|
 |
Biphenyl-2,3-diol 1,2-dioxygenase. Chain: a. Synonym: 23ohbp oxygenase, 2,3-dihydroxybiphenyl dioxygenase, dhbd. Engineered: yes
|
|
Source:
|
 |
Burkholderia xenovorans. Organism_taxid: 266265. Strain: lb400. Expressed in: pseudomonas putida. Expression_system_taxid: 303. Other_details: burkholderia sp. Strain lb400 has been reclassifed. Prior publications may refer to this organism as pseudomonas sp. Strain lb400 or burkholderia cepacia strain lb400. See m.G.Fain, j.D.Haddock, current microbiol. (2001) 42:269-73
|
|
Biol. unit:
|
 |
 Octamer (from PDB file)
Octamer (from PDB file)
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.203
|
R-free:
|
0.220
|
|
|
Authors:
|
 |
S.Dai,J.T.Bolin
|
Key ref:

|
 |
S.Dai
et al.
(2002).
Identification and analysis of a bottleneck in PCB biodegradation.
Nat Struct Biol,
9,
934-939.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
24-Apr-02
|
Release date:
|
27-Nov-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P47228
(BPHC_PARXL) -
Biphenyl-2,3-diol 1,2-dioxygenase from Paraburkholderia xenovorans (strain LB400)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
298 a.a.
287 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.39
- biphenyl-2,3-diol 1,2-dioxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
biphenyl-2,3-diol + O2 = 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate + H+
|
 |
 |
 |
 |
 |
biphenyl-2,3-diol
Bound ligand (Het Group name = )
matches with 87.50% similarity
|
+
|
 O2
O2
|
=
|
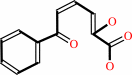 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate
2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mn(2+) or Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Biol
9:934-939
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Identification and analysis of a bottleneck in PCB biodegradation.
|
|
S.Dai,
F.H.Vaillancourt,
H.Maaroufi,
N.M.Drouin,
D.B.Neau,
V.Snieckus,
J.T.Bolin,
L.D.Eltis.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The microbial degradation of polychlorinated biphenyls (PCBs) provides the
potential to destroy these widespread, toxic and persistent environmental
pollutants. For example, the four-step upper bph pathway transforms some of the
more than 100 different PCBs found in commercial mixtures and is being
engineered for more effective PCB degradation. In the critical third step of
this pathway, 2,3-dihydroxybiphenyl (DHB) 1,2-dioxygenase (DHBD; EC 1.13.11.39)
catalyzes aromatic ring cleavage. Here we demonstrate that ortho-chlorinated PCB
metabolites strongly inhibit DHBD, promote its suicide inactivation and
interfere with the degradation of other compounds. For example, k(cat)(app) for
2',6'-diCl DHB was reduced by a factor of approximately 7,000 relative to DHB,
and it bound with sufficient affinity to competitively inhibit DHB cleavage at
nanomolar concentrations. Crystal structures of two complexes of DHBD with
ortho-chlorinated metabolites at 1.7 A resolution reveal an explanation for
these phenomena, which have important implications for bioremediation strategies.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
|
|

|
| |
The above figure is
reprinted
by permission from Macmillan Publishers Ltd:
Nat Struct Biol
(2002,
9,
934-939)
copyright 2002.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.C.Yam,
S.Okamoto,
J.N.Roberts,
and
L.D.Eltis
(2011).
Adventures in Rhodococcus - from steroids to explosives.
|
| |
Can J Microbiol,
57,
155-168.
|
 |
|
|
|
|
 |
K.C.Yam,
I.D'Angelo,
R.Kalscheuer,
H.Zhu,
J.X.Wang,
V.Snieckus,
L.H.Ly,
P.J.Converse,
W.R.Jacobs,
N.Strynadka,
and
L.D.Eltis
(2009).
Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis.
|
| |
PLoS Pathog,
5,
e1000344.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.F.Pacios,
V.M.Campos,
I.Merino,
and
L.Gómez
(2009).
Structures and thermodynamics of biphenyl dihydrodiol stereoisomers and their metabolites in the enzymatic degradation of arene xenobiotics.
|
| |
J Comput Chem,
30,
2420-2432.
|
 |
|
|
|
|
 |
K.Furukawa,
and
H.Fujihara
(2008).
Microbial degradation of polychlorinated biphenyls: biochemical and molecular features.
|
| |
J Biosci Bioeng,
105,
433-449.
|
 |
|
|
|
|
 |
S.A.Adebusoye,
F.W.Picardal,
M.O.Ilori,
and
O.O.Amund
(2008).
Evidence of aerobic utilization of di-ortho-substituted trichlorobiphenyls as growth substrates by Pseudomonas sp. SA-6 and Ralstonia sp. SA-4.
|
| |
Environ Microbiol,
10,
1165-1174.
|
 |
|
|
|
|
 |
S.A.Adebusoye,
F.W.Picardal,
M.O.Ilori,
O.O.Amund,
C.Fuqua,
and
N.Grindle
(2007).
Growth on dichlorobiphenyls with chlorine substitution on each ring by bacteria isolated from contaminated African soils.
|
| |
Appl Microbiol Biotechnol,
74,
484-492.
|
 |
|
|
|
|
 |
S.Bhowmik,
G.P.Horsman,
J.T.Bolin,
and
L.D.Eltis
(2007).
The molecular basis for inhibition of BphD, a C-C bond hydrolase involved in polychlorinated biphenyls degradation: large 3-substituents prevent tautomerization.
|
| |
J Biol Chem,
282,
36377-36385.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Feliciano,
S.Xu,
X.Guan,
H.J.Lehmler,
L.G.Bachas,
and
S.Daunert
(2006).
ClcR-based biosensing system in the detection of cis-dihydroxylated (chloro-)biphenyls.
|
| |
Anal Bioanal Chem,
385,
807-813.
|
 |
|
|
|
|
 |
P.D.Fortin,
G.P.Horsman,
H.M.Yang,
and
L.D.Eltis
(2006).
A glutathione S-transferase catalyzes the dehalogenation of inhibitory metabolites of polychlorinated biphenyls.
|
| |
J Bacteriol,
188,
4424-4430.
|
 |
|
|
|
|
 |
V.J.Denef,
J.A.Klappenbach,
M.A.Patrauchan,
C.Florizone,
J.L.Rodrigues,
T.V.Tsoi,
W.Verstraete,
L.D.Eltis,
and
J.M.Tiedje
(2006).
Genetic and genomic insights into the role of benzoate-catabolic pathway redundancy in Burkholderia xenovorans LB400.
|
| |
Appl Environ Microbiol,
72,
585-595.
|
 |
|
|
|
|
 |
D.H.Pieper
(2005).
Aerobic degradation of polychlorinated biphenyls.
|
| |
Appl Microbiol Biotechnol,
67,
170-191.
|
 |
|
|
|
|
 |
J.L.Pellequer,
S.W.Chen,
Y.S.Keum,
A.E.Karu,
Q.X.Li,
and
V.A.Roberts
(2005).
Structural basis for preferential binding of non-ortho-substituted polychlorinated biphenyls by the monoclonal antibody S2B1.
|
| |
J Mol Recognit,
18,
282-294.
|
 |
|
|
|
|
 |
M.L.Neidig,
and
E.I.Solomon
(2005).
Structure-function correlations in oxygen activating non-heme iron enzymes.
|
| |
Chem Commun (Camb),
(),
5843-5863.
|
 |
|
|
|
|
 |
P.D.Fortin,
A.T.Lo,
M.A.Haro,
S.R.Kaschabek,
W.Reineke,
and
L.D.Eltis
(2005).
Evolutionarily divergent extradiol dioxygenases possess higher specificities for polychlorinated biphenyl metabolites.
|
| |
J Bacteriol,
187,
415-421.
|
 |
|
|
|
|
 |
P.D.Fortin,
I.MacPherson,
D.B.Neau,
J.T.Bolin,
and
L.D.Eltis
(2005).
Directed evolution of a ring-cleaving dioxygenase for polychlorinated biphenyl degradation.
|
| |
J Biol Chem,
280,
42307-42314.
|
 |
|
|
|
|
 |
S.Tillmann,
C.Strömpl,
K.N.Timmis,
and
W.R.Abraham
(2005).
Stable isotope probing reveals the dominant role of Burkholderia species in aerobic degradation of PCBs.
|
| |
FEMS Microbiol Ecol,
52,
207-217.
|
 |
|
|
|
|
 |
V.J.Denef,
M.A.Patrauchan,
C.Florizone,
J.Park,
T.V.Tsoi,
W.Verstraete,
J.M.Tiedje,
and
L.D.Eltis
(2005).
Growth substrate- and phase-specific expression of biphenyl, benzoate, and C1 metabolic pathways in Burkholderia xenovorans LB400.
|
| |
J Bacteriol,
187,
7996-8005.
|
 |
|
|
|
|
 |
V.J.Denef,
J.Park,
T.V.Tsoi,
J.M.Rouillard,
H.Zhang,
J.A.Wibbenmeyer,
W.Verstraete,
E.Gulari,
S.A.Hashsham,
and
J.M.Tiedje
(2004).
Biphenyl and benzoate metabolism in a genomic context: outlining genome-wide metabolic networks in Burkholderia xenovorans LB400.
|
| |
Appl Environ Microbiol,
70,
4961-4970.
|
 |
|
|
|
|
 |
D.B.McKay,
M.Prucha,
W.Reineke,
K.N.Timmis,
and
D.H.Pieper
(2003).
Substrate specificity and expression of three 2,3-dihydroxybiphenyl 1,2-dioxygenases from Rhodococcus globerulus strain P6.
|
| |
J Bacteriol,
185,
2944-2951.
|
 |
|
|
|
|
 |
F.H.Vaillancourt,
M.A.Haro,
N.M.Drouin,
Z.Karim,
H.Maaroufi,
and
L.D.Eltis
(2003).
Characterization of extradiol dioxygenases from a polychlorinated biphenyl-degrading strain that possess higher specificities for chlorinated metabolites.
|
| |
J Bacteriol,
185,
1253-1260.
|
 |
|
|
|
|
 |
K.Furukawa
(2003).
'Super bugs' for bioremediation.
|
| |
Trends Biotechnol,
21,
187-190.
|
 |
|
|
|
|
 |
N.Gilmartin,
D.Ryan,
O.Sherlock,
and
D.Dowling
(2003).
BphK shows dechlorination activity against 4-chlorobenzoate, an end product of bph-promoted degradation of PCBs.
|
| |
FEMS Microbiol Lett,
222,
251-255.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

