 |
PDBsum entry 1l1d

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1l1d
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.8.4.11
- peptide-methionine (S)-S-oxide reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
L-methionyl-[protein] + [thioredoxin]-disulfide + H2O = L-methionyl- (S)-S-oxide-[protein] + [thioredoxin]-dithiol
|
|
2.
|
[thioredoxin]-disulfide + L-methionine + H2O = L-methionine (S)-S- oxide + [thioredoxin]-dithiol
|
|
 |
 |
 |
 |
 |
L-methionyl-[protein]
|
+
|
[thioredoxin]-disulfide
|
+
|
H2O
|
=
|
L-methionyl- (S)-S-oxide-[protein]
|
+
|
[thioredoxin]-dithiol
|
|
 |
 |
 |
 |
 |
[thioredoxin]-disulfide
|
+
|
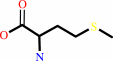 L-methionine
L-methionine
|
+
|
H2O
|
=
|
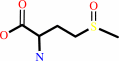 L-methionine (S)-S- oxide
L-methionine (S)-S- oxide
|
+
|
[thioredoxin]-dithiol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.8.4.12
- peptide-methionine (R)-S-oxide reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-methionyl-[protein] + [thioredoxin]-disulfide + H2O = L-methionyl-(R)- S-oxide-[protein] + [thioredoxin]-dithiol
|
 |
 |
 |
 |
 |
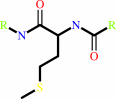 Peptide-L-methionine
Peptide-L-methionine
|
+
|
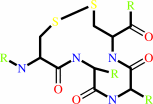 thioredoxin disulfide
thioredoxin disulfide
|
+
|
H(2)O
|
=
|
peptide-L- methionine (R)-S-oxide
|
+
|
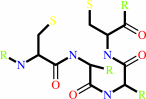 thioredoxin
thioredoxin
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Se(2+); Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Biol
9:348-352
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
The mirrored methionine sulfoxide reductases of Neisseria gonorrhoeae pilB.
|
|
W.T.Lowther,
H.Weissbach,
F.Etienne,
N.Brot,
B.W.Matthews.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Methionine sulfoxide reductases (Msr) protect against oxidative damage that can
contribute to cell death. The tandem Msr domains (MsrA and MsrB) of the pilB
protein from Neisseria gonorrhoeae each reduce different epimeric forms of
methionine sulfoxide. The overall fold of the MsrB domain revealed by the 1.85 A
crystal structure shows no resemblance to the previously determined MsrA
structures from other organisms. Despite the lack of homology, the active sites
show approximate mirror symmetry. In each case, conserved amino acid motifs
mediate the stereo-specific recognition and reduction of the substrate. Unlike
the MsrA domain, the MsrB domain activates the cysteine or selenocysteine
nucleophile through a unique Cys-Arg-Asp/Glu catalytic triad. The collapse of
the reaction intermediate most likely results in the formation of a sulfenic or
selenenic acid moiety. Regeneration of the active site occurs through a series
of thiol-disulfide exchange steps involving another active site Cys residue and
thioredoxin. These observations have broad implications for modular catalysis,
antibiotic drug design and continuing longevity studies in mammals.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Mirror-like relationship between the active sites of
MsrB and MsrA domains. a, Model for the interactions of
Met-R(O) (orange carbon atoms) with the active site of the pilB
MsrB domain (this work). b, Model for the interaction of
Met-S(O) with the active site of bovine MsrA^16. In each case,
only small adjustments to the rotamer angle of the presumed
nucleophilic Cys residue, Cys 495 or Cys 72, were required to
establish a feasible interaction (  2.1
Å) with the sulfur atom of the sulfoxide moiety (dashed
yellow lines). The substrates were also docked in a manner to
place the oxygen and sulfur atoms at the axial positions of the
putative trigonal-bipyramidal intermediate of the reaction (Fig.
4). The MsrA domain of pilB presumably shows interactions
similar to those here because the active site residues are
conserved and the overall sequence identity is 40% (ref. 15). 2.1
Å) with the sulfur atom of the sulfoxide moiety (dashed
yellow lines). The substrates were also docked in a manner to
place the oxygen and sulfur atoms at the axial positions of the
putative trigonal-bipyramidal intermediate of the reaction (Fig.
4). The MsrA domain of pilB presumably shows interactions
similar to those here because the active site residues are
conserved and the overall sequence identity is 40% (ref. 15).
|
 |
Figure 4.
Figure 4. Proposed reaction mechanism for MsrB catalysis. The
active site residues and the solvent molecule, W 1, are
indicated in bold. The nucleophilic attack by Cys 495 (I)
results in a trigonal-bipyramidal intermediate (II). Collapse of
the intermediate leads to the formation of a sulfenic acid
derivative of Cys 495 and the release of methionine (III). The
analogous derivative within the MsrB homolog SelX probably
contains a selenenic acid modification (Fig. 2a). A series of
thiol-disulfide exchange reactions (III and IV) and the action
of reducing thiols, either dithiothreitol (DTT) in vitro or the
thioredoxin-thioredoxin reductase system (TR/TRR) with its
associated cofactor NADPH in vivo, return the active site to the
fully reduced state. Whether the addition of the proton to the
oxygen atom of the sulfoxide moiety occurs between steps I and
II or steps II and III, as shown, remains to be determined.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Struct Biol
(2002,
9,
348-352)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.C.Lee,
and
V.N.Gladyshev
(2011).
The biological significance of methionine sulfoxide stereochemistry.
|
| |
Free Radic Biol Med,
50,
221-227.
|
 |
|
|
|
|
 |
M.Carella,
J.Becher,
O.Ohlenschläger,
R.Ramachandran,
K.H.Gührs,
G.Wellenreuther,
W.Meyer-Klaucke,
S.H.Heinemann,
and
M.Görlach
(2011).
Structure-function relationship in an archaebacterial methionine sulphoxide reductase B.
|
| |
Mol Microbiol,
79,
342-358.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Iglesias-Baena,
S.Barranco-Medina,
A.Lázaro-Payo,
F.J.López-Jaramillo,
F.Sevilla,
and
J.J.Lázaro
(2010).
Characterization of plant sulfiredoxin and role of sulphinic form of 2-Cys peroxiredoxin.
|
| |
J Exp Bot,
61,
1509-1521.
|
 |
|
|
|
|
 |
M.Carella,
O.Ohlenschläger,
R.Ramachandran,
and
M.Görlach
(2010).
1H, 13C and 15N resonance assignment of a zinc-binding methionine sulfoxide reductase type-B from the thermophilic archeabacterium Methanothermobacter thermoautotrophicus.
|
| |
Biomol NMR Assign,
4,
93-95.
|
 |
|
|
|
|
 |
N.Ugarte,
I.Petropoulos,
and
B.Friguet
(2010).
Oxidized mitochondrial protein degradation and repair in aging and oxidative stress.
|
| |
Antioxid Redox Signal,
13,
539-549.
|
 |
|
|
|
|
 |
D.T.Le,
B.C.Lee,
S.M.Marino,
Y.Zhang,
D.E.Fomenko,
A.Kaya,
E.Hacioglu,
G.H.Kwak,
A.Koc,
H.Y.Kim,
and
V.N.Gladyshev
(2009).
Functional Analysis of Free Methionine-R-sulfoxide Reductase from Saccharomyces cerevisiae.
|
| |
J Biol Chem,
284,
4354-4364.
|
 |
|
|
|
|
 |
J.P.Jacquot,
H.Eklund,
N.Rouhier,
and
P.Schürmann
(2009).
Structural and evolutionary aspects of thioredoxin reductases in photosynthetic organisms.
|
| |
Trends Plant Sci,
14,
336-343.
|
 |
|
|
|
|
 |
R.J.Hondal
(2009).
Using chemical approaches to study selenoproteins-focus on thioredoxin reductases.
|
| |
Biochim Biophys Acta,
1790,
1501-1512.
|
 |
|
|
|
|
 |
T.Takenawa,
A.Yokota,
M.Oda,
H.Takahashi,
and
M.Iwakura
(2009).
Protein oxidation during long storage: identification of the oxidation sites in dihydrofolate reductase from Escherichia coli through LC-MS and fragment studies.
|
| |
J Biochem,
145,
517-523.
|
 |
|
|
|
|
 |
Y.K.Kim,
Y.J.Shin,
W.H.Lee,
H.Y.Kim,
and
K.Y.Hwang
(2009).
Structural and kinetic analysis of an MsrA-MsrB fusion protein from Streptococcus pneumoniae.
|
| |
Mol Microbiol,
72,
699-709.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.T.Le,
X.Liang,
D.E.Fomenko,
A.S.Raza,
C.K.Chong,
B.A.Carlson,
D.L.Hatfield,
and
V.N.Gladyshev
(2008).
Analysis of methionine/selenomethionine oxidation and methionine sulfoxide reductase function using methionine-rich proteins and antibodies against their oxidized forms.
|
| |
Biochemistry,
47,
6685-6694.
|
 |
|
|
|
|
 |
F.R.Salsbury,
S.T.Knutson,
L.B.Poole,
and
J.S.Fetrow
(2008).
Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid.
|
| |
Protein Sci,
17,
299-312.
|
 |
|
|
|
|
 |
X.H.Zhang,
and
H.Weissbach
(2008).
Origin and evolution of the protein-repairing enzymes methionine sulphoxide reductases.
|
| |
Biol Rev Camb Philos Soc,
83,
249-257.
|
 |
|
|
|
|
 |
A.De Luca,
P.Sacchetta,
M.Nieddu,
C.Di Ilio,
and
B.Favaloro
(2007).
Important roles of multiple Sp1 binding sites and epigenetic modifications in the regulation of the methionine sulfoxide reductase B1 (MsrB1) promoter.
|
| |
BMC Mol Biol,
8,
39.
|
 |
|
|
|
|
 |
B.Chen,
L.M.Markillie,
Y.Xiong,
M.U.Mayer,
and
T.C.Squier
(2007).
Increased catalytic efficiency following gene fusion of bifunctional methionine sulfoxide reductase enzymes from Shewanella oneidensis.
|
| |
Biochemistry,
46,
14153-14161.
|
 |
|
|
|
|
 |
F.Neiers,
S.Sonkaria,
A.Olry,
S.Boschi-Muller,
and
G.Branlant
(2007).
Characterization of the amino acids from Neisseria meningitidis methionine sulfoxide reductase B involved in the chemical catalysis and substrate specificity of the reductase step.
|
| |
J Biol Chem,
282,
32397-32405.
|
 |
|
|
|
|
 |
L.Delaye,
A.Becerra,
L.Orgel,
and
A.Lazcano
(2007).
Molecular evolution of peptide methionine sulfoxide reductases (MsrA and MsrB): on the early development of a mechanism that protects against oxidative damage.
|
| |
J Mol Evol,
64,
15-32.
|
 |
|
|
|
|
 |
N.Rouhier,
B.Kauffmann,
F.Tete-Favier,
P.Palladino,
P.Gans,
G.Branlant,
J.P.Jacquot,
and
S.Boschi-Muller
(2007).
Functional and structural aspects of poplar cytosolic and plastidial type a methionine sulfoxide reductases.
|
| |
J Biol Chem,
282,
3367-3378.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.J.Sasindran,
S.Saikolappan,
and
S.Dhandayuthapani
(2007).
Methionine sulfoxide reductases and virulence of bacterial pathogens.
|
| |
Future Microbiol,
2,
619-630.
|
 |
|
|
|
|
 |
Z.Lin,
L.C.Johnson,
H.Weissbach,
N.Brot,
M.O.Lively,
and
W.T.Lowther
(2007).
Free methionine-(R)-sulfoxide reductase from Escherichia coli reveals a new GAF domain function.
|
| |
Proc Natl Acad Sci U S A,
104,
9597-9602.
|
 |
|
|
|
|
 |
D.Sagher,
D.Brunell,
J.F.Hejtmancik,
M.Kantorow,
N.Brot,
and
H.Weissbach
(2006).
Thionein can serve as a reducing agent for the methionine sulfoxide reductases.
|
| |
Proc Natl Acad Sci U S A,
103,
8656-8661.
|
 |
|
|
|
|
 |
D.Sagher,
D.Brunell,
N.Brot,
B.L.Vallee,
and
H.Weissbach
(2006).
Selenocompounds can serve as oxidoreductants with the methionine sulfoxide reductase enzymes.
|
| |
J Biol Chem,
281,
31184-31187.
|
 |
|
|
|
|
 |
H.Y.Kim,
D.E.Fomenko,
Y.E.Yoon,
and
V.N.Gladyshev
(2006).
Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases.
|
| |
Biochemistry,
45,
13697-13704.
|
 |
|
|
|
|
 |
M.B.Cannon,
and
S.J.Remington
(2006).
Re-engineering redox-sensitive green fluorescent protein for improved response rate.
|
| |
Protein Sci,
15,
45-57.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Brot,
J.F.Collet,
L.C.Johnson,
T.J.Jönsson,
H.Weissbach,
and
W.T.Lowther
(2006).
The thioredoxin domain of Neisseria gonorrhoeae PilB can use electrons from DsbD to reduce downstream methionine sulfoxide reductases.
|
| |
J Biol Chem,
281,
32668-32675.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Alamuri,
and
R.J.Maier
(2006).
Methionine sulfoxide reductase in Helicobacter pylori: interaction with methionine-rich proteins and stress-induced expression.
|
| |
J Bacteriol,
188,
5839-5850.
|
 |
|
|
|
|
 |
A.Olry,
S.Boschi-Muller,
H.Yu,
D.Burnel,
and
G.Branlant
(2005).
Insights into the role of the metal binding site in methionine-R-sulfoxide reductases B.
|
| |
Protein Sci,
14,
2828-2837.
|
 |
|
|
|
|
 |
B.Ezraty,
J.Bos,
F.Barras,
and
L.Aussel
(2005).
Methionine sulfoxide reduction and assimilation in Escherichia coli: new role for the biotin sulfoxide reductase BisC.
|
| |
J Bacteriol,
187,
231-237.
|
 |
|
|
|
|
 |
H.Y.Kim,
and
V.N.Gladyshev
(2005).
Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases.
|
| |
PLoS Biol,
3,
e375.
|
 |
|
|
|
|
 |
J.Walter,
P.Chagnaud,
G.W.Tannock,
D.M.Loach,
F.Dal Bello,
H.F.Jenkinson,
W.P.Hammes,
and
C.Hertel
(2005).
A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut.
|
| |
Appl Environ Microbiol,
71,
979-986.
|
 |
|
|
|
|
 |
J.Wu,
F.Neiers,
S.Boschi-Muller,
and
G.Branlant
(2005).
The N-terminal domain of PILB from Neisseria meningitidis is a disulfide reductase that can recycle methionine sulfoxide reductases.
|
| |
J Biol Chem,
280,
12344-12350.
|
 |
|
|
|
|
 |
R.Bentley
(2005).
Role of sulfur chirality in the chemical processes of biology.
|
| |
Chem Soc Rev,
34,
609-624.
|
 |
|
|
|
|
 |
F.Neiers,
A.Kriznik,
S.Boschi-Muller,
and
G.Branlant
(2004).
Evidence for a new sub-class of methionine sulfoxide reductases B with an alternative thioredoxin recognition signature.
|
| |
J Biol Chem,
279,
42462-42468.
|
 |
|
|
|
|
 |
H.Y.Kim,
and
V.N.Gladyshev
(2004).
Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases.
|
| |
Mol Biol Cell,
15,
1055-1064.
|
 |
|
|
|
|
 |
L.B.Poole,
P.A.Karplus,
and
A.Claiborne
(2004).
Protein sulfenic acids in redox signaling.
|
| |
Annu Rev Pharmacol Toxicol,
44,
325-347.
|
 |
|
|
|
|
 |
O.Yermolaieva,
R.Xu,
C.Schinstock,
N.Brot,
H.Weissbach,
S.H.Heinemann,
and
T.Hoshi
(2004).
Methionine sulfoxide reductase A protects neuronal cells against brief hypoxia/reoxygenation.
|
| |
Proc Natl Acad Sci U S A,
101,
1159-1164.
|
 |
|
|
|
|
 |
P.F.Mugford,
S.M.Lait,
B.A.Keay,
and
R.J.Kazlauskas
(2004).
Enantiocomplementary enzymatic resolution of the chiral auxiliary: cis,cis-6-(2,2-dimethylpropanamido)spiro[4.4]nonan-1-ol and the molecular basis for the high enantioselectivity of subtilisin Carlsberg.
|
| |
Chembiochem,
5,
980-987.
|
 |
|
|
|
|
 |
T.Douglas,
D.S.Daniel,
B.K.Parida,
C.Jagannath,
and
S.Dhandayuthapani
(2004).
Methionine sulfoxide reductase A (MsrA) deficiency affects the survival of Mycobacterium smegmatis within macrophages.
|
| |
J Bacteriol,
186,
3590-3598.
|
 |
|
|
|
|
 |
A.B.Taylor,
D.M.Benglis,
S.Dhandayuthapani,
and
P.J.Hart
(2003).
Structure of Mycobacterium tuberculosis methionine sulfoxide reductase A in complex with protein-bound methionine.
|
| |
J Bacteriol,
185,
4119-4126.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.R.Stadtman,
J.Moskovitz,
and
R.L.Levine
(2003).
Oxidation of methionine residues of proteins: biological consequences.
|
| |
Antioxid Redox Signal,
5,
577-582.
|
 |
|
|
|
|
 |
M.Antoine,
S.Boschi-Muller,
and
G.Branlant
(2003).
Kinetic characterization of the chemical steps involved in the catalytic mechanism of methionine sulfoxide reductase A from Neisseria meningitidis.
|
| |
J Biol Chem,
278,
45352-45357.
|
 |
|
|
|
|
 |
E.P.Skaar,
D.M.Tobiason,
J.Quick,
R.C.Judd,
H.Weissbach,
F.Etienne,
N.Brot,
and
H.S.Seifert
(2002).
The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species.
|
| |
Proc Natl Acad Sci U S A,
99,
10108-10113.
|
 |
|
|
|
|
 |
R.A.Kumar,
A.Koc,
R.L.Cerny,
and
V.N.Gladyshev
(2002).
Reaction mechanism, evolutionary analysis, and role of zinc in Drosophila methionine-R-sulfoxide reductase.
|
| |
J Biol Chem,
277,
37527-37535.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

