 |
PDBsum entry 1jc4

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.1.99.1
- methylmalonyl-CoA epimerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-methylmalonyl-CoA = (S)-methylmalonyl-CoA
|
 |
 |
 |
 |
 |
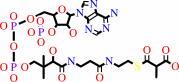 (R)-methylmalonyl-CoA
(R)-methylmalonyl-CoA
|
=
|
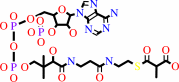 (S)-methylmalonyl-CoA
(S)-methylmalonyl-CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
9:637-646
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of methylmalonyl-coenzyme A epimerase from P. shermanii: a novel enzymatic function on an ancient metal binding scaffold.
|
|
A.A.McCarthy,
H.M.Baker,
S.C.Shewry,
M.L.Patchett,
E.N.Baker.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Methylmalonyl-CoA epimerase (MMCE) is an essential enzyme in the
breakdown of odd-numbered fatty acids and of the amino acids valine, isoleucine,
and methionine. Present in many bacteria and in animals, it catalyzes the
conversion of (2R)-methylmalonyl-CoA to (2S)-methylmalonyl-CoA, the substrate
for the B12-dependent enzyme, methylmalonyl-CoA mutase. Defects in this pathway
can result in severe acidosis and cause damage to the central nervous system in
humans. RESULTS: The crystal structure of MMCE from Propionibacterium shermanii
has been determined at 2.0 A resolution. The MMCE monomer is folded into two
tandem betaalphabetabetabeta modules that pack edge-to-edge to generate an
8-stranded beta sheet. Two monomers then pack back-to-back to create a tightly
associated dimer. In each monomer, the beta sheet curves around to create a deep
cleft, in the floor of which His12, Gln65, His91, and Glu141 provide a binding
site for a divalent metal ion, as shown by the binding of Co2+. Modeling
2-methylmalonate into the active site identifies two glutamate residues as the
likely essential bases for the epimerization reaction. CONCLUSIONS: The
betaalphabetabetabeta modules of MMCE correspond with those found in several
other proteins, including bleomycin resistance protein, glyoxalase I, and a
family of extradiol dioxygenases. Differences in connectivity are consistent
with the evolution of these very different proteins from a common precursor by
mechanisms of gene duplication and domain swapping. The metal binding residues
also align precisely, and striking structural similarities between MMCE and
glyoxalase I suggest common mechanisms in their respective epimerization and
isomerization reactions.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 4.
Figure 4. The Metal Binding Site in MMCE(a) The difference
electron density peak for the bound Co2+ ion is shown, in
coordinating distance of the side chains of His12, Gln65, His91,
and Glu141. Virtually no movement of these coordinating groups
occurs on metal complexation to the apo-protein.(b) The metal
sites of MMCE and GLO are superimposed using only the Ca atoms
of the two proteins for superposition. For MMCE, the polypeptide
backbone is in gray, with side chains and the Co2+ ion in blue;
for GLO, the polypeptide backbone is in black, with side chains
in red (monomer A) and gold (monomer B), and the Zn2+ ion in red

|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2001,
9,
637-646)
copyright 2001.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Xu
(2010).
Enhancing MAD F(A) data for substructure determination.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
945-949.
|
 |
|
|
|
|
 |
J.Zheng,
C.A.Taylor,
S.K.Piasecki,
and
A.T.Keatinge-Clay
(2010).
Structural and functional analysis of A-type ketoreductases from the amphotericin modular polyketide synthase.
|
| |
Structure,
18,
913-922.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Morar,
and
G.D.Wright
(2010).
The genomic enzymology of antibiotic resistance.
|
| |
Annu Rev Genet,
44,
25-51.
|
 |
|
|
|
|
 |
L.Shi,
P.Gao,
X.X.Yan,
and
D.C.Liang
(2009).
Crystal structure of a putative methylmalonyl-coenzyme a epimerase from Thermoanaerobacter tengcongensis at 2.0 A resolution.
|
| |
Proteins,
77,
994-999.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.Wu,
P.M.Flatt,
H.Xu,
and
T.Mahmud
(2009).
Biosynthetic Gene Cluster of Cetoniacytone A, an Unusual Aminocyclitol from the Endosymbiotic Bacterium Actinomyces sp. Lu 9419.
|
| |
Chembiochem,
10,
304-314.
|
 |
|
|
|
|
 |
C.Andreini,
I.Bertini,
G.Cavallaro,
G.L.Holliday,
and
J.M.Thornton
(2008).
Metal ions in biological catalysis: from enzyme databases to general principles.
|
| |
J Biol Inorg Chem,
13,
1205-1218.
|
 |
|
|
|
|
 |
C.Dumas,
and
A.van der Lee
(2008).
Macromolecular structure solution by charge flipping.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
864-873.
|
 |
|
|
|
|
 |
H.Xu,
and
C.M.Weeks
(2008).
Rapid and automated substructure solution by Shake-and-Bake.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
172-177.
|
 |
|
|
|
|
 |
I.Mulako,
J.M.Farrant,
H.Collett,
and
N.Illing
(2008).
Expression of Xhdsi-1VOC, a novel member of the vicinal oxygen chelate (VOC) metalloenzyme superfamily, is up-regulated in leaves and roots during desiccation in the resurrection plant Xerophyta humilis (Bak) Dur and Schinz.
|
| |
J Exp Bot,
59,
3885-3901.
|
 |
|
|
|
|
 |
N.Sukdeo,
and
J.F.Honek
(2008).
Microbial glyoxalase enzymes: metalloenzymes controlling cellular levels of methylglyoxal.
|
| |
Drug Metabol Drug Interact,
23,
29-50.
|
 |
|
|
|
|
 |
T.J.Erb,
J.Rétey,
G.Fuchs,
and
B.E.Alber
(2008).
Ethylmalonyl-CoA mutase from Rhodobacter sphaeroides defines a new subclade of coenzyme B12-dependent acyl-CoA mutases.
|
| |
J Biol Chem,
283,
32283-32293.
|
 |
|
|
|
|
 |
F.Ferrè,
G.Ausiello,
A.Zanzoni,
and
M.Helmer-Citterich
(2005).
Functional annotation by identification of local surface similarities: a novel tool for structural genomics.
|
| |
BMC Bioinformatics,
6,
194.
|
 |
|
|
|
|
 |
F.H.Vaillancourt,
E.Yeh,
D.A.Vosburg,
S.E.O'Connor,
and
C.T.Walsh
(2005).
Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis.
|
| |
Nature,
436,
1191-1194.
|
 |
|
|
|
|
 |
H.Xu,
C.M.Weeks,
and
H.A.Hauptman
(2005).
Optimizing statistical Shake-and-Bake for Se-atom substructure determination.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
976-981.
|
 |
|
|
|
|
 |
J.Kühnl,
T.Bobik,
J.B.Procter,
C.Burmeister,
J.Höppner,
I.Wilde,
K.Lüersen,
A.E.Torda,
R.D.Walter,
and
E.Liebau
(2005).
Functional analysis of the methylmalonyl-CoA epimerase from Caenorhabditis elegans.
|
| |
FEBS J,
272,
1465-1477.
|
 |
|
|
|
|
 |
R.G.Zhang,
N.Duke,
R.Laskowski,
E.Evdokimova,
T.Skarina,
A.Edwards,
A.Joachimiak,
and
A.Savchenko
(2003).
Conserved protein YecM from Escherichia coli shows structural homology to metal-binding isomerases and oxygenases.
|
| |
Proteins,
51,
311-314.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.W.Martin,
Z.Dauter,
Y.Devedjiev,
P.Sheffield,
F.Jelen,
M.He,
D.H.Sherman,
J.Otlewski,
Z.S.Derewenda,
and
U.Derewenda
(2002).
Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
|
| |
Structure,
10,
933-942.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

