
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chain B:
E.C.3.2.2.6
- ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
NAD+ + H2O = ADP-D-ribose + nicotinamide + H+
|
 |
 |
 |
 |
 |
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
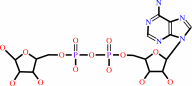 ADP-D-ribose
ADP-D-ribose
|
+
|
 nicotinamide
nicotinamide
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nature
386:190-194
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the type-I interleukin-1 receptor complexed with interleukin-1beta.
|
|
G.P.Vigers,
L.J.Anderson,
P.Caffes,
B.J.Brandhuber.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Interleukin-1 (IL-1) is an important mediator of inflammatory disease. The IL-1
family currently consists of two agonists, IL-1alpha and IL-1beta, and one
antagonist, IL-1ra. Each of these molecules binds to the type I IL-1 receptor
(IL1R). The binding of IL-1alpha or IL-1beta to IL1R is an early step in IL-1
signal transduction and blocking this interaction may therefore be a useful
target for the development of new drugs. Here we report the three-dimensional
structure of IL-1beta bound to the extracellular domain of IL1R (s-IL1R) at 2.5
A resolution. IL-1beta binds to s-IL1R with a 1:1 stoichiometry. The crystal
structure shows that s-IL1R consists of three immunoglobulin-like domains which
wrap around IL-1beta in a manner distinct from the structures of previously
described cytokine-receptor complexes. The two receptor-binding regions on
IL-1beta identified by site-directed mutagenesis both contact the receptor: one
binds to the first two domains of the receptor, while the other binds
exclusively to the third domain.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Thomas,
J.F.Bazan,
and
K.C.Garcia
(2012).
Structure of the activating IL-1 receptor signaling complex.
|
| |
Nat Struct Mol Biol,
19,
455-457.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.M.Owyang,
H.Issafras,
J.Corbin,
K.Ahluwalia,
P.Larsen,
E.Pongo,
M.Handa,
A.H.Horwitz,
M.K.Roell,
M.Haak-Frendscho,
and
L.Masat
(2011).
XOMA 052, a potent, high-affinity monoclonal antibody for the treatment of IL-1β-mediated diseases.
|
| |
MAbs,
3,
49-60.
|
 |
|
|
|
|
 |
N.Boutard,
S.Turcotte,
K.Beauregard,
C.Quiniou,
S.Chemtob,
and
W.D.Lubell
(2011).
Examination of the active secondary structure of the peptide 101.10, an allosteric modulator of the interleukin-1 receptor, by positional scanning using β-amino γ-lactams.
|
| |
J Pept Sci,
17,
288-296.
|
 |
|
|
|
|
 |
X.Zhang,
P.Angkasekwinai,
C.Dong,
and
H.Tang
(2011).
Structure and function of interleukin-17 family cytokines.
|
| |
Protein Cell,
2,
26-40.
|
 |
|
|
|
|
 |
D.Wang,
S.Zhang,
L.Li,
X.Liu,
K.Mei,
and
X.Wang
(2010).
Structural insights into the assembly and activation of IL-1β with its receptors.
|
| |
Nat Immunol,
11,
905-911.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.E.Sims
(2010).
Accessory to inflammation.
|
| |
Nat Immunol,
11,
883-885.
|
 |
|
|
|
|
 |
J.H.Lee,
L.C.Wang,
H.H.Yu,
Y.T.Lin,
Y.H.Yang,
and
B.L.Chiang
(2010).
Type I IL-1 receptor (IL-1RI) as potential new therapeutic target for bronchial asthma.
|
| |
Mediators Inflamm,
2010,
567351.
|
 |
|
|
|
|
 |
X.Zhang,
F.Shephard,
H.B.Kim,
I.R.Palmer,
S.McHarg,
G.J.Fowler,
L.A.O'Neill,
E.Kiss-Toth,
and
E.E.Qwarnstrom
(2010).
TILRR, a novel IL-1RI Co-receptor, potentiates MyD88 recruitment to control Ras-dependent amplification of NF-kappaB.
|
| |
J Biol Chem,
285,
7222-7232.
|
 |
|
|
|
|
 |
A.Lingel,
T.M.Weiss,
M.Niebuhr,
B.Pan,
B.A.Appleton,
C.Wiesmann,
J.F.Bazan,
and
W.J.Fairbrother
(2009).
Structure of IL-33 and its interaction with the ST2 and IL-1RAcP receptors--insight into heterotrimeric IL-1 signaling complexes.
|
| |
Structure,
17,
1398-1410.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.W.Schmid,
M.A.Lynch,
and
C.E.Herron
(2009).
The effects of IL-1 receptor antagonist on beta amyloid mediated depression of LTP in the rat CA1 in vivo.
|
| |
Hippocampus,
19,
670-676.
|
 |
|
|
|
|
 |
K.L.Hailey,
S.Li,
M.D.Andersen,
M.Roy,
V.L.Woods,
and
P.A.Jennings
(2009).
Pro-interleukin (IL)-1beta shares a core region of stability as compared with mature IL-1beta while maintaining a distinctly different configurational landscape: a comparative hydrogen/deuterium exchange mass spectrometry study.
|
| |
J Biol Chem,
284,
26137-26148.
|
 |
|
|
|
|
 |
M.A.Argiriadi,
T.Xiang,
C.Wu,
T.Ghayur,
and
D.W.Borhani
(2009).
Unusual water-mediated antigenic recognition of the proinflammatory cytokine interleukin-18.
|
| |
J Biol Chem,
284,
24478-24489.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.M.Luheshi,
N.J.Rothwell,
and
D.Brough
(2009).
Dual functionality of interleukin-1 family cytokines: implications for anti-interleukin-1 therapy.
|
| |
Br J Pharmacol,
157,
1318-1329.
|
 |
|
|
|
|
 |
W.B.Wang,
J.M.Fan,
X.L.Zhang,
J.Xu,
and
W.Yao
(2009).
Serial expression analysis of liver regeneration-related genes in rat regenerating liver.
|
| |
Mol Biotechnol,
43,
221-231.
|
 |
|
|
|
|
 |
B.Krumm,
X.Meng,
Y.Li,
Y.Xiang,
and
J.Deng
(2008).
Structural basis for antagonism of human interleukin 18 by poxvirus interleukin 18-binding protein.
|
| |
Proc Natl Acad Sci U S A,
105,
20711-20715.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Gosavi,
P.C.Whitford,
P.A.Jennings,
and
J.N.Onuchic
(2008).
Extracting function from a beta-trefoil folding motif.
|
| |
Proc Natl Acad Sci U S A,
105,
10384-10389.
|
 |
|
|
|
|
 |
G.Vergoten,
and
J.P.Zanetta
(2007).
Structural differences between the putative carbohydrate-recognition domains of human IL-1 alpha, IL-1 beta and IL-1 receptor antagonist obtained by in silico modeling.
|
| |
Glycoconj J,
24,
183-193.
|
 |
|
|
|
|
 |
S.Ali,
M.Huber,
C.Kollewe,
S.C.Bischoff,
W.Falk,
and
M.U.Martin
(2007).
IL-1 receptor accessory protein is essential for IL-33-induced activation of T lymphocytes and mast cells.
|
| |
Proc Natl Acad Sci U S A,
104,
18660-18665.
|
 |
|
|
|
|
 |
S.Yuzawa,
Y.Opatowsky,
Z.Zhang,
V.Mandiyan,
I.Lax,
and
J.Schlessinger
(2007).
Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor.
|
| |
Cell,
130,
323-334.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Hänel,
T.Stangler,
M.Stoldt,
and
D.Willbold
(2006).
Solution structure of the X4 protein coded by the SARS related coronavirus reveals an immunoglobulin like fold and suggests a binding activity to integrin I domains.
|
| |
J Biomed Sci,
13,
281-293.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.A.Langer,
E.C.Cutrone,
and
S.Kotenko
(2004).
The Class II cytokine receptor (CRF2) family: overview and patterns of receptor-ligand interactions.
|
| |
Cytokine Growth Factor Rev,
15,
33-48.
|
 |
|
|
|
|
 |
N.Lozovaya,
and
A.D.Miller
(2003).
Chemical neuroimmunology: health in a nutshell bidirectional communication between immune and stress (limbic-hypothalamic-pituitary-adrenal) systems.
|
| |
Chembiochem,
4,
466-484.
|
 |
|
|
|
|
 |
Z.Kato,
J.Jee,
H.Shikano,
M.Mishima,
I.Ohki,
H.Ohnishi,
A.Li,
K.Hashimoto,
E.Matsukuma,
K.Omoya,
Y.Yamamoto,
T.Yoneda,
T.Hara,
N.Kondo,
and
M.Shirakawa
(2003).
The structure and binding mode of interleukin-18.
|
| |
Nat Struct Biol,
10,
966-971.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.E.Smith,
R.R.Ketchem,
H.Moore,
Z.Anderson,
B.R.Renshaw,
D.J.Friend,
and
J.E.Sims
(2002).
A single amino acid difference between human and monkey interleukin (IL)-1beta dictates effective binding to soluble type II IL-1 receptor.
|
| |
J Biol Chem,
277,
47619-47625.
|
 |
|
|
|
|
 |
J.E.Sims
(2002).
IL-1 and IL-18 receptors, and their extended family.
|
| |
Curr Opin Immunol,
14,
117-122.
|
 |
|
|
|
|
 |
J.R.Hea,
S.Bino,
G.W.Roberts,
J.G.Raynes,
and
A.D.Miller
(2002).
Mechanistic investigation into complementary (antisense) peptide mini-receptor inhibitors of cytokine interleukin-1.
|
| |
Chembiochem,
3,
76-85.
|
 |
|
|
|
|
 |
S.Bird,
J.Zou,
T.Wang,
B.Munday,
C.Cunningham,
and
C.J.Secombes
(2002).
Evolution of interleukin-1beta.
|
| |
Cytokine Growth Factor Rev,
13,
483-502.
|
 |
|
|
|
|
 |
S.H.Kim,
T.Azam,
D.Novick,
D.Y.Yoon,
L.L.Reznikov,
P.Bufler,
M.Rubinstein,
and
C.A.Dinarello
(2002).
Identification of amino acid residues critical for biological activity in human interleukin-18.
|
| |
J Biol Chem,
277,
10998-11003.
|
 |
|
|
|
|
 |
W.J.Kao,
Y.Liu,
R.Gundloori,
J.Li,
D.Lee,
N.Einerson,
J.Burmania,
and
K.Stevens
(2002).
Engineering endogenous inflammatory cells as delivery vehicles.
|
| |
J Control Release,
78,
219-233.
|
 |
|
|
|
|
 |
B.Spörri,
M.Bickel,
D.Dobbelaere,
J.Machado,
and
D.Lottaz
(2001).
Soluble interleukin-1 receptor--reverse signaling in innate immunoregulation.
|
| |
Cytokine Growth Factor Rev,
12,
27-32.
|
 |
|
|
|
|
 |
E.Dunn,
J.E.Sims,
M.J.Nicklin,
and
L.A.O'Neill
(2001).
Annotating genes with potential roles in the immune system: six new members of the IL-1 family.
|
| |
Trends Immunol,
22,
533-536.
|
 |
|
|
|
|
 |
J.T.Bensen,
P.A.Dawson,
J.C.Mychaleckyj,
and
D.W.Bowden
(2001).
Identification of a novel human cytokine gene in the interleukin gene cluster on chromosome 2q12-14.
|
| |
J Interferon Cytokine Res,
21,
899-904.
|
 |
|
|
|
|
 |
S.H.Kim,
T.Azam,
D.Y.Yoon,
L.L.Reznikov,
D.Novick,
M.Rubinstein,
and
C.A.Dinarello
(2001).
Site-specific mutations in the mature form of human IL-18 with enhanced biological activity and decreased neutralization by IL-18 binding protein.
|
| |
Proc Natl Acad Sci U S A,
98,
3304-3309.
|
 |
|
|
|
|
 |
V.M.Abramov,
A.M.Vasiliev,
R.N.Vasilenko,
N.L.Kulikova,
I.V.Kosarev,
V.S.Khlebnikov,
A.T.Ishchenko,
S.MacIntyre,
J.R.Gillespie,
R.Khurana,
T.Korpela,
A.L.Fink,
and
V.N.Uversky
(2001).
Structural and functional similarity between Yersinia pestis capsular protein Caf1 and human interleukin-1 beta.
|
| |
Biochemistry,
40,
6076-6084.
|
 |
|
|
|
|
 |
B.A.Chrunyk,
M.H.Rosner,
Y.Cong,
A.S.McColl,
I.G.Otterness,
and
G.O.Daumy
(2000).
Inhibiting protein-protein interactions: a model for antagonist design.
|
| |
Biochemistry,
39,
7092-7099.
|
 |
|
|
|
|
 |
C.A.Dinarello
(2000).
Targeting interleukin 18 with interleukin 18 binding protein.
|
| |
Ann Rheum Dis,
59,
i17-i20.
|
 |
|
|
|
|
 |
D.J.Stauber,
A.D.DiGabriele,
and
W.A.Hendrickson
(2000).
Structural interactions of fibroblast growth factor receptor with its ligands.
|
| |
Proc Natl Acad Sci U S A,
97,
49-54.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.J.Thiel,
M.H.le Du,
R.L.Walter,
A.D'Arcy,
C.Chène,
M.Fountoulakis,
G.Garotta,
F.K.Winkler,
and
S.E.Ealick
(2000).
Observation of an unexpected third receptor molecule in the crystal structure of human interferon-gamma receptor complex.
|
| |
Structure,
8,
927-936.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.C.Deller,
and
E.Yvonne Jones
(2000).
Cell surface receptors.
|
| |
Curr Opin Struct Biol,
10,
213-219.
|
 |
|
|
|
|
 |
S.H.Kim,
M.Eisenstein,
L.Reznikov,
G.Fantuzzi,
D.Novick,
M.Rubinstein,
and
C.A.Dinarello
(2000).
Structural requirements of six naturally occurring isoforms of the IL-18 binding protein to inhibit IL-18.
|
| |
Proc Natl Acad Sci U S A,
97,
1190-1195.
|
 |
|
|
|
|
 |
S.Kumar,
P.C.McDonnell,
R.Lehr,
L.Tierney,
M.N.Tzimas,
D.E.Griswold,
E.A.Capper,
R.Tal-Singer,
G.I.Wells,
M.L.Doyle,
and
P.R.Young
(2000).
Identification and initial characterization of four novel members of the interleukin-1 family.
|
| |
J Biol Chem,
275,
10308-10314.
|
 |
|
|
|
|
 |
V.P.Smith,
and
A.Alcami
(2000).
Expression of secreted cytokine and chemokine inhibitors by ectromelia virus.
|
| |
J Virol,
74,
8460-8471.
|
 |
|
|
|
|
 |
F.Quatra,
M.R.Colonna,
and
A.Catalano
(1999).
The role of interleukin-1alpha and collagenase in chronic wounds.
|
| |
Plast Reconstr Surg,
104,
1205-1206.
|
 |
|
|
|
|
 |
G.Venkataraman,
R.Raman,
V.Sasisekharan,
and
R.Sasisekharan
(1999).
Molecular characteristics of fibroblast growth factor-fibroblast growth factor receptor-heparin-like glycosaminoglycan complex.
|
| |
Proc Natl Acad Sci U S A,
96,
3658-3663.
|
 |
|
|
|
|
 |
M.T.Huhtala,
O.T.Pentikäinen,
and
M.S.Johnson
(1999).
A dimeric ternary complex of FGFR [correction of FGFR1], heparin and FGF-1 leads to an 'electrostatic sandwich' model for heparin binding.
|
| |
Structure,
7,
699-709.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Secombes,
J.Zou,
G.Daniels,
C.Cunningham,
A.Koussounadis,
and
G.Kemp
(1998).
Rainbow trout cytokine and cytokine receptor genes.
|
| |
Immunol Rev,
166,
333-340.
|
 |
|
|
|
|
 |
L.A.Goldman,
E.C.Cutrone,
A.Dang,
X.Hao,
J.K.Lim,
and
J.A.Langer
(1998).
Mapping human interferon-alpha (IFN-alpha 2) binding determinants of the type I interferon receptor subunit IFNAR-1 with human/bovine IFNAR-1 chimeras.
|
| |
Biochemistry,
37,
13003-13010.
|
 |
|
|
|
|
 |
P.E.Auron
(1998).
The interleukin 1 receptor: ligand interactions and signal transduction.
|
| |
Cytokine Growth Factor Rev,
9,
221-237.
|
 |
|
|
|
|
 |
S.C.Garman,
J.P.Kinet,
and
T.S.Jardetzky
(1998).
Crystal structure of the human high-affinity IgE receptor.
|
| |
Cell,
95,
951-961.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.P.Arend,
M.Malyak,
C.J.Guthridge,
and
C.Gabay
(1998).
Interleukin-1 receptor antagonist: role in biology.
|
| |
Annu Rev Immunol,
16,
27-55.
|
 |
|
|
|
|
 |
C.A.Dinarello
(1997).
Interleukin-1.
|
| |
Cytokine Growth Factor Rev,
8,
253-265.
|
 |
|
|
|
|
 |
R.Singh,
S.Huang,
T.Guth,
M.Konieczkowski,
and
J.R.Sedor
(1997).
Cytosolic domain of the type I interleukin-1 receptor spontaneously recruits signaling molecules to activate a proinflammatory gene.
|
| |
J Clin Invest,
100,
419-428.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

