 |
PDBsum entry 1idt

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1idt
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.5.1.34
- 6,7-dihydropteridine reductase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Biopterin Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
5,6,7,8-tetrahydropteridine + NADP+ = 6,7-dihydropteridine + NADPH + H+
|
|
2.
|
5,6,7,8-tetrahydropteridine + NAD+ = 6,7-dihydropteridine + NADH + H+
|
|
 |
 |
 |
 |
 |
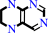 5,6,7,8-tetrahydropteridine
5,6,7,8-tetrahydropteridine
|
+
|
 NADP(+)
NADP(+)
|
=
|
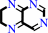 6,7-dihydropteridine
6,7-dihydropteridine
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
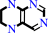 5,6,7,8-tetrahydropteridine
5,6,7,8-tetrahydropteridine
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 41.51% similarity
|
=
|
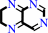 6,7-dihydropteridine
6,7-dihydropteridine
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Med Chem
46:4009-4020
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Studies on the nitroreductase prodrug-activating system. Crystal structures of complexes with the inhibitor dicoumarol and dinitrobenzamide prodrugs and of the enzyme active form.
|
|
E.Johansson,
G.N.Parkinson,
W.A.Denny,
S.Neidle.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The E. coli nitroreductase enzyme (NTR) has been widely used in suicide gene
therapy (GDEPT and ADEPT) applications as a activating enzyme for nitroaromatic
prodrugs of the dinitrobenzamide class. NTR has been previously shown to be a
homodimeric enzyme with two active sites. We present here the crystal structures
of the reduced form of NTR and its complexes with the inhibitor dicoumarol and
three dinitrobenzamide prodrugs. Comparison of the structures of the oxidized
and reduced forms of the native enzyme shows that the principal structural
changes occur in the FMN cofactor and indicate that the enzyme itself is a
relatively rigid structure that primarily provides a rigid structural framework
on which hydride transfer occurs. The aziridinyldinitrobenzamide prodrug CB 1954
binds in nonidentical ways in both of the two active sites of the homodimeric
enzyme, employing both hydrophobic and (in active site B) a direct H-bond
contact to the side chain of Lys14. In active site A the 2-nitro group stacks
above the FMN, and in active site B the 4-nitro group does, explaining why
reduction of either nitro group is observed. In contrast, the larger mustard
group of the dinitrobenzamide mustard compound SN 23862 forces the prodrug to
bind at both active sites with only the 2-nitro group able to participate in
hydride transfer from the FMN, explaining why only the 2-hydroxylamine reduction
product is observed. In each site, the nitro groups of the prodrug make direct
H-bond contacts with the enzyme; in active Site A the 2-nitro to Ser40 and the
4-nitro to Asn71, while in active Site B the 2-nitro contacts the main chain
nitrogen of Thr41 and the 4-nitro group the Lys14 side chain. The related
amide-substituted mustard SN 27217 binds in a broadly similar fashion, but with
the larger amide group substituent able to reach and contact the side chain of
Arg107, further restricting the prodrug conformations in the binding site. The
inhibitor dicoumarol appears to bind primarily by pi-stacking interactions and
hydrophobic contacts, with no conformational changes in the enzyme. One of the
hydroxycoumarin subunits stacks above the plane of the FMN via pi-overlap with
the isoalloxazine ring, penetrating deep into the groove, with the other less
well-defined. These studies suggest guidelines for further prodrug design.
Steric bulk (e.g., mustard rather than aziridine) on the ring can limit the
possible binding orientations, and the reducible nitro group must be located
para to the mustard. Substitution on the carboxamide side chain still allows the
prodrugs to bind, but also limits their orientation in the binding site.
Finally, modulating substrate specificity by alteration of the structure of the
enzyme rather than the prodrug might usefully focus on modifying the Phe124
residue and those surrounding it.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Manina,
M.Bellinzoni,
M.R.Pasca,
J.Neres,
A.Milano,
A.L.Ribeiro,
S.Buroni,
H.Skovierová,
P.Dianišková,
K.Mikušová,
J.Marák,
V.Makarov,
D.Giganti,
A.Haouz,
A.P.Lucarelli,
G.Degiacomi,
A.Piazza,
L.R.Chiarelli,
E.De Rossi,
E.Salina,
S.T.Cole,
P.M.Alzari,
and
G.Riccardi
(2010).
Biological and structural characterization of the Mycobacterium smegmatis nitroreductase NfnB, and its role in benzothiazinone resistance.
|
| |
Mol Microbiol,
77,
1172-1185.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.Rong,
W.Du,
Y.Wang,
and
L.Lu
(2010).
Carbon supported Pt colloid as effective catalyst for selective hydrogenation of nitroarenes to arylhydroxylamines.
|
| |
Chem Commun (Camb),
46,
1559-1561.
|
 |
|
|
|
|
 |
A.F.Tavares,
L.S.Nobre,
A.M.Melo,
and
L.M.Saraiva
(2009).
A novel nitroreductase of Staphylococcus aureus with S-nitrosoglutathione reductase activity.
|
| |
J Bacteriol,
191,
3403-3406.
|
 |
|
|
|
|
 |
S.R.Thomas,
P.M.McTamney,
J.M.Adler,
N.Laronde-Leblanc,
and
S.E.Rokita
(2009).
Crystal structure of iodotyrosine deiodinase, a novel flavoprotein responsible for iodide salvage in thyroid glands.
|
| |
J Biol Chem,
284,
19659-19667.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.D.Roldán,
E.Pérez-Reinado,
F.Castillo,
and
C.Moreno-Vivián
(2008).
Reduction of polynitroaromatic compounds: the bacterial nitroreductases.
|
| |
FEMS Microbiol Rev,
32,
474-500.
|
 |
|
|
|
|
 |
D.C.Singleton,
D.Li,
S.Y.Bai,
S.P.Syddall,
J.B.Smaill,
Y.Shen,
W.A.Denny,
W.R.Wilson,
and
A.V.Patterson
(2007).
The nitroreductase prodrug SN 28343 enhances the potency of systemically administered armed oncolytic adenovirus ONYX-411(NTR).
|
| |
Cancer Gene Ther,
14,
953-967.
|
 |
|
|
|
|
 |
H.Iwaki,
T.Muraki,
S.Ishihara,
Y.Hasegawa,
K.N.Rankin,
T.Sulea,
J.Boyd,
and
P.C.Lau
(2007).
Characterization of a pseudomonad 2-nitrobenzoate nitroreductase and its catabolic pathway-associated 2-hydroxylaminobenzoate mutase and a chemoreceptor involved in 2-nitrobenzoate chemotaxis.
|
| |
J Bacteriol,
189,
3502-3514.
|
 |
|
|
|
|
 |
H.Pisharath,
J.M.Rhee,
M.A.Swanson,
S.D.Leach,
and
M.J.Parsons
(2007).
Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase.
|
| |
Mech Dev,
124,
218-229.
|
 |
|
|
|
|
 |
K.Takeda,
M.Iizuka,
T.Watanabe,
J.Nakagawa,
S.Kawasaki,
and
Y.Niimura
(2007).
Synechocystis DrgA protein functioning as nitroreductase and ferric reductase is capable of catalyzing the Fenton reaction.
|
| |
FEBS J,
274,
1318-1327.
|
 |
|
|
|
|
 |
M.M.AbuKhader,
J.Heap,
C.I.De Matteis,
S.W.Doughty,
N.Minton,
and
M.Paoli
(2007).
Crystallization and preliminary X-ray characterization of the Bacillus amyloliquefaciens YwrO enzyme.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
746-750.
|
 |
|
|
|
|
 |
W.R.Wilson,
K.O.Hicks,
S.M.Pullen,
D.M.Ferry,
N.A.Helsby,
and
A.V.Patterson
(2007).
Bystander effects of bioreductive drugs: potential for exploiting pathological tumor hypoxia with dinitrobenzamide mustards.
|
| |
Radiat Res,
167,
625-636.
|
 |
|
|
|
|
 |
Z.C.Symons,
and
N.C.Bruce
(2006).
Bacterial pathways for degradation of nitroaromatics.
|
| |
Nat Prod Rep,
23,
845-850.
|
 |
|
|
|
|
 |
P.R.Race,
A.L.Lovering,
R.M.Green,
A.Ossor,
S.A.White,
P.F.Searle,
C.J.Wrighton,
and
E.I.Hyde
(2005).
Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
|
| |
J Biol Chem,
280,
13256-13264.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Kutty,
and
G.N.Bennett
(2005).
Biochemical characterization of trinitrotoluene transforming oxygen-insensitive nitroreductases from Clostridium acetobutylicum ATCC 824.
|
| |
Arch Microbiol,
184,
158-167.
|
 |
|
|
|
|
 |
P.F.Searle,
M.J.Chen,
L.Hu,
P.R.Race,
A.L.Lovering,
J.I.Grove,
C.Guise,
M.Jaberipour,
N.D.James,
V.Mautner,
L.S.Young,
D.J.Kerr,
A.Mountain,
S.A.White,
and
E.I.Hyde
(2004).
Nitroreductase: a prodrug-activating enzyme for cancer gene therapy.
|
| |
Clin Exp Pharmacol Physiol,
31,
811-816.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

