 |
PDBsum entry 1i5e

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.9
- uracil phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
UMP + diphosphate = 5-phospho-alpha-D-ribose 1-diphosphate + uracil
|
 |
 |
 |
 |
 |
 UMP
UMP
|
+
|
diphosphate
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
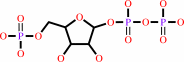 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
+
|
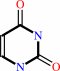 uracil
uracil
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
58:936-945
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of product-bound Bacillus caldolyticus uracil phosphoribosyltransferase confirms ordered sequential substrate binding.
|
|
A.Kadziola,
J.Neuhard,
S.Larsen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Uracil phosphoribosyltransferase (UPRTase) is part of the salvage pathway that
leads to the biosynthesis of UMP. It catalyzes the formation of UMP and
pyrophosphate from uracil and alpha-D-5-phosphoribosyl-1-pyrophosphate. Unlike
enzymes in the de novo synthesis of UMP, UPRTases have only been found in lower
organisms and are therefore potential targets for the development of new
antibiotics. UPRTase from Bacillus caldolyticus has been crystallized and the
structure has been determined by isomorphous replacement and refined to 3.0 A
resolution. UPRTase from B. caldolyticus forms a dimer with the active sites
pointing away from each other. A long arm from each subunit wraps around the
other subunit, contributing half of the dimer interface. The monomer adopts the
phosphoribosyltransferase type I fold, with a small C-terminal hood defining the
uracil-binding site. The structure contains a well defined UMP molecule in the
active site. The binding of UMP involves two sequence segments that are highly
conserved among UPRTases. The first segment, Asp131-Ser139, contains the
PRPP-binding consensus sequence motif known from other type I
phosphoribosyltransferases and binds the ribose-5'-phosphate part of UMP. The
second segment, Tyr193-Ala201, which is specific for uracil
phosphoribosyltransferases, binds the uracil part of UMP through backbone
contacts, partly mediated by a water molecule. Modelling of a PRPP-enzyme
complex reveals that uracil can be activated to its tautomeric enol form by the
complex. This is consistent with kinetic data, which display ordered sequential
binding of substrates, with PRPP binding first. Based on this observation, a
reaction mechanism is proposed.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 7.
Figure 7 Stereoview of a hypothetical enzyme-substrate complex.
The substrate molecules uracil and PRPP are shown (black bonds)
superposed onto the actual enzyme-product complex (white bonds)
for which the structure was determined. The ribose-5'-phosphate
part common to substrate and product has grey bonds.
|
 |
Figure 8.
Figure 8 Proposed catalytic mechanism for uracil
phosphoribosyltransferase: when PRPP is bound to the enzyme and
uracil enters the active site (Fig. 7-), uracil is stabilized as
the enol tautomer. When in the enol form, uracil enters deeper
into the active site and the shown electron translocation takes
place, completing the reaction.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2002,
58,
936-945)
copyright 2002.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.L.Turnbough,
and
R.L.Switzer
(2008).
Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors.
|
| |
Microbiol Mol Biol Rev,
72,
266.
|
 |
|
|
|
|
 |
K.F.Jensen,
S.Arent,
S.Larsen,
and
L.Schack
(2005).
Allosteric properties of the GTP activated and CTP inhibited uracil phosphoribosyltransferase from the thermoacidophilic archaeon Sulfolobus solfataricus.
|
| |
FEBS J,
272,
1440-1453.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

