 |
PDBsum entry 1hm5

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.3.1.9
- glucose-6-phosphate isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
alpha-D-glucose 6-phosphate = beta-D-fructose 6-phosphate
|
 |
 |
 |
 |
 |
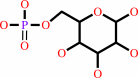 alpha-D-glucose 6-phosphate
alpha-D-glucose 6-phosphate
|
=
|
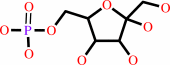 beta-D-fructose 6-phosphate
beta-D-fructose 6-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
323:77-84
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Conformational changes in phosphoglucose isomerase induced by ligand binding.
|
|
D.Arsenieva,
C.J.Jeffery.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Phosphoglucose isomerase (PGI; EC 5.3.1.9) is the second enzyme in glycolysis,
where it catalyzes the isomerization of D-glucose-6-phosphate to
D-fructose-6-phosphate. It is the same protein as autocrine motility factor,
differentiation and maturation mediator, and neuroleukin. Here, we report a new
X-ray crystal structure of rabbit PGI (rPGI) without ligands bound in its active
site. The structure was solved at 1.8A resolution by isomorphous phasing with a
previously solved X-ray crystal structure of the rPGI dimer containing
6-phosphogluconate in its active site. Comparison of the new structure to
previously reported structures enables identification of conformational changes
that occur during binding of substrate or inhibitor molecules. Ligand binding
causes an induced fit of regions containing amino acid residues 209-215, 245-259
and 385-389. This conformational change differs from the change previously
reported to occur between the ring-opening and isomerization steps, in which the
helix containing residues 513-521 moves toward the bound substrate. Differences
between the liganded and unliganded structures are limited to the region within
and close to the active-site pocket.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Conformational changes in the active site of rPGI
during catalysis. The unliganded structure is shown in blue. The
enzyme in complex with F6P is shown in green. The complex with
5PAH is shown in red. Ordered water molecules are indicated by a
number accompanied by the letter W. (a) Active site of
unliganded rPGI. An electron density map calculated with
coefficients |2F[o] -F[c]| is shown with a 1s contour level
(gray). The active site is filled with ordered water molecules.
(b) Movements in the active site induced by substrate binding. A
partial alpha carbon trace indicates regions that move upon F6P
binding. (Alpha carbon atoms of amino acid residues 1-555 were
superposed, only the regions that move are shown.) The F6P
ligand is in dark green, and water molecules displaced by F6P
are labeled in blue. (c) Conformational changes in the active
site between the ring opening and isomerization steps. An alpha
carbon trace indicates the helix containing amino acid residues
513-521 that shifts after the ring opening step but before the
isomerization step. That movement is different from the movement
of regions induced by F6P binding. This Figure and Figure 2 were
made using the program BOBSCRIPT.42
|
 |
Figure 3.
Figure 3. Surface shape and potential of rPGI with and
without bound ligands. The molecular surface shape and charge
distribution do not change significantly upon ligand binding,
except in the region of the active site. The protein is oriented
similarly in all four panels. The surface is colored according
to charge: positively charged groups are blue, negatively
charged groups are red, and uncharged groups are gray. In (a),
unliganded rPGI, the black rectangle indicates the active-site
pocket. The active site is filled in (b) the rPGI/F6P complex,
(c) the rPGI/5PAH complex, and (d) the rPGI/6PGA complex.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2002,
323,
77-84)
copyright 2002.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.K.Wierenga,
E.G.Kapetaniou,
and
R.Venkatesan
(2010).
Triosephosphate isomerase: a highly evolved biocatalyst.
|
| |
Cell Mol Life Sci,
67,
3961-3982.
|
 |
|
|
|
|
 |
M.Fairbank,
P.St-Pierre,
and
I.R.Nabi
(2009).
The complex biology of autocrine motility factor/phosphoglucose isomerase (AMF/PGI) and its receptor, the gp78/AMFR E3 ubiquitin ligase.
|
| |
Mol Biosyst,
5,
793-801.
|
 |
|
|
|
|
 |
C.Roux,
N.Gresh,
L.E.Perera,
J.P.Piquemal,
and
L.Salmon
(2007).
Binding of 5-phospho-D-arabinonohydroxamate and 5-phospho-D-arabinonate inhibitors to zinc phosphomannose isomerase from Candida albicans studied by polarizable molecular mechanics and quantum mechanics.
|
| |
J Comput Chem,
28,
938-957.
|
 |
|
|
|
|
 |
C.W.Wheat,
W.B.Watt,
D.D.Pollock,
and
P.M.Schulte
(2006).
From DNA to fitness differences: sequences and structures of adaptive variants of Colias phosphoglucose isomerase (PGI).
|
| |
Mol Biol Evol,
23,
499-512.
|
 |
|
|
|
|
 |
J.H.Lee,
and
C.J.Jeffery
(2005).
The crystal structure of rabbit phosphoglucose isomerase complexed with D-sorbitol-6-phosphate, an analog of the open chain form of D-glucose-6-phosphate.
|
| |
Protein Sci,
14,
727-734.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Hansen,
B.Schlichting,
M.Felgendreher,
and
P.Schönheit
(2005).
Cupin-type phosphoglucose isomerases (Cupin-PGIs) constitute a novel metal-dependent PGI family representing a convergent line of PGI evolution.
|
| |
J Bacteriol,
187,
1621-1631.
|
 |
|
|
|
|
 |
A.T.Cordeiro,
P.A.Michels,
L.F.Delboni,
and
O.H.Thiemann
(2004).
The crystal structure of glucose-6-phosphate isomerase from Leishmania mexicana reveals novel active site features.
|
| |
Eur J Biochem,
271,
2765-2772.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.K.Swan,
T.Hansen,
P.Schönheit,
and
C.Davies
(2004).
A novel phosphoglucose isomerase (PGI)/phosphomannose isomerase from the crenarchaeon Pyrobaculum aerophilum is a member of the PGI superfamily: structural evidence at 1.16-A resolution.
|
| |
J Biol Chem,
279,
39838-39845.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Sanishvili,
R.Wu,
D.E.Kim,
J.D.Watson,
F.Collart,
and
A.Joachimiak
(2004).
Crystal structure of Bacillus subtilis YckF: structural and functional evolution.
|
| |
J Struct Biol,
148,
98.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Hansen,
D.Wendorff,
and
P.Schönheit
(2004).
Bifunctional phosphoglucose/phosphomannose isomerases from the Archaea Aeropyrum pernix and Thermoplasma acidophilum constitute a novel enzyme family within the phosphoglucose isomerase superfamily.
|
| |
J Biol Chem,
279,
2262-2272.
|
 |
|
|
|
|
 |
C.Davies,
H.Muirhead,
and
J.Chirgwin
(2003).
The structure of human phosphoglucose isomerase complexed with a transition-state analogue.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1111-1113.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.K.Swan,
J.T.Solomons,
C.C.Beeson,
T.Hansen,
P.Schönheit,
and
C.Davies
(2003).
Structural evidence for a hydride transfer mechanism of catalysis in phosphoglucose isomerase from Pyrococcus furiosus.
|
| |
J Biol Chem,
278,
47261-47268.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

