 |
PDBsum entry 1gsn

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1gsn
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Human glutathione reductase modified by dinitrosoglutathione
|
|
Structure:
|
 |
Glutathione reductase. Chain: a. Engineered: yes. Other_details: mixed disulfide between c58 and glutathione (gsh 1030) sulfenic acid group in cea63, cea234, cea284, cea423
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.207
|
R-free:
|
0.249
|
|
|
Authors:
|
 |
K.Becker,S.N.Savvides,M.Keese,R.H.Schirmer,P.A.Karplus
|
|
Key ref:
|
 |
K.Becker
et al.
(1998).
Enzyme inactivation through sulfhydryl oxidation by physiologic NO-carriers.
Nat Struct Biol,
5,
267-271.
PubMed id:
|
 |
|
Date:
|
 |
|
21-Feb-98
|
Release date:
|
27-May-98
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00390
(GSHR_HUMAN) -
Glutathione reductase, mitochondrial from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
522 a.a.
461 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 4 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.8.1.7
- glutathione-disulfide reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 glutathione + NADP+ = glutathione disulfide + NADPH + H+
|
 |
 |
 |
 |
 |
2
×
glutathione
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NADP(+)
NADP(+)
|
=
|
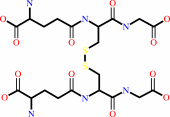 glutathione disulfide
glutathione disulfide
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nat Struct Biol
5:267-271
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Enzyme inactivation through sulfhydryl oxidation by physiologic NO-carriers.
|
|
K.Becker,
S.N.Savvides,
M.Keese,
R.H.Schirmer,
P.A.Karplus.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Nitric oxide (NO) is a pluripotent regulatory molecule, yet the molecular
mechanisms by which it exerts its effects are largely unknown. Few physiologic
target molecules of NO have been identified, and even for these, the
modifications caused by NO remain uncharacterized. Human glutathione reductase
(hGR), a central enzyme of cellular antioxidant defense, is inhibited by
S-nitrosoglutathione (GSNO) and by diglutathionyl-dinitroso-iron (DNIC-[GSH]2),
two in vivo transport forms of NO. Here, crystal structures of hGR inactivated
by GSNO and DNIC-[GSH]2 at 1.7 A resolution provide the first picture of enzyme
inactivation by NO-carriers: in GSNO-modified hGR, the active site residue Cys
63 is oxidized to an unusually stable cysteine sulfenic acid (R-SOH), whereas
modification with DNIC-[GSH]2 oxidizes Cys 63 to a cysteine sulfinic acid
(R-SO2H). Our results illustrate that various forms of NO can mediate distinct
chemistry, and that sulfhydryl oxidation must be considered as a major mechanism
of NO action.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.S.Rehder,
and
C.R.Borges
(2010).
Possibilities and pitfalls in quantifying the extent of cysteine sulfenic acid modification of specific proteins within complex biofluids.
|
| |
BMC Biochem,
11,
25.
|
 |
|
|
|
|
 |
E.T.Chouchani,
T.R.Hurd,
S.M.Nadtochiy,
P.S.Brookes,
I.M.Fearnley,
K.S.Lilley,
R.A.Smith,
and
M.P.Murphy
(2010).
Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation.
|
| |
Biochem J,
430,
49-59.
|
 |
|
|
|
|
 |
G.Rai,
A.A.Sayed,
W.A.Lea,
H.F.Luecke,
H.Chakrapani,
S.Prast-Nielsen,
A.Jadhav,
W.Leister,
M.Shen,
J.Inglese,
C.P.Austin,
L.Keefer,
E.S.Arnér,
A.Simeonov,
D.J.Maloney,
D.L.Williams,
and
C.J.Thomas
(2009).
Structure mechanism insights and the role of nitric oxide donation guide the development of oxadiazole-2-oxides as therapeutic agents against schistosomiasis.
|
| |
J Med Chem,
52,
6474-6483.
|
 |
|
|
|
|
 |
F.R.Salsbury,
S.T.Knutson,
L.B.Poole,
and
J.S.Fetrow
(2008).
Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid.
|
| |
Protein Sci,
17,
299-312.
|
 |
|
|
|
|
 |
P.S.Smith-Pearson,
M.Kooshki,
D.R.Spitz,
L.B.Poole,
W.Zhao,
and
M.E.Robbins
(2008).
Decreasing peroxiredoxin II expression decreases glutathione, alters cell cycle distribution, and sensitizes glioma cells to ionizing radiation and H(2)O(2).
|
| |
Free Radic Biol Med,
45,
1178-1189.
|
 |
|
|
|
|
 |
A.Diet,
K.Abbas,
C.Bouton,
B.Guillon,
F.Tomasello,
S.Fourquet,
M.B.Toledano,
and
J.C.Drapier
(2007).
Regulation of peroxiredoxins by nitric oxide in immunostimulated macrophages.
|
| |
J Biol Chem,
282,
36199-36205.
|
 |
|
|
|
|
 |
G.Wang,
C.Strang,
P.J.Pfaffinger,
and
M.Covarrubias
(2007).
Zn2+-dependent redox switch in the intracellular T1-T1 interface of a Kv channel.
|
| |
J Biol Chem,
282,
13637-13647.
|
 |
|
|
|
|
 |
J.Z.Pedersen,
F.De Maria,
P.Turella,
G.Federici,
M.Mattei,
R.Fabrini,
K.F.Dawood,
M.Massimi,
A.M.Caccuri,
and
G.Ricci
(2007).
Glutathione transferases sequester toxic dinitrosyl-iron complexes in cells. A protection mechanism against excess nitric oxide.
|
| |
J Biol Chem,
282,
6364-6371.
|
 |
|
|
|
|
 |
L.Stella,
V.Pallottini,
S.Moreno,
S.Leoni,
F.De Maria,
P.Turella,
G.Federici,
R.Fabrini,
K.F.Dawood,
M.L.Bello,
J.Z.Pedersen,
and
G.Ricci
(2007).
Electrostatic association of glutathione transferase to the nuclear membrane. Evidence of an enzyme defense barrier at the nuclear envelope.
|
| |
J Biol Chem,
282,
6372-6379.
|
 |
|
|
|
|
 |
S.Mocellin,
V.Bronte,
and
D.Nitti
(2007).
Nitric oxide, a double edged sword in cancer biology: searching for therapeutic opportunities.
|
| |
Med Res Rev,
27,
317-352.
|
 |
|
|
|
|
 |
C.C.Dahm,
K.Moore,
and
M.P.Murphy
(2006).
Persistent S-nitrosation of complex I and other mitochondrial membrane proteins by S-nitrosothiols but not nitric oxide or peroxynitrite: implications for the interaction of nitric oxide with mitochondria.
|
| |
J Biol Chem,
281,
10056-10065.
|
 |
|
|
|
|
 |
R.Singh,
M.A.White,
K.V.Ramana,
J.M.Petrash,
S.J.Watowich,
A.Bhatnagar,
and
S.K.Srivastava
(2006).
Structure of a glutathione conjugate bound to the active site of aldose reductase.
|
| |
Proteins,
64,
101-110.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Urig,
K.Fritz-Wolf,
R.Réau,
C.Herold-Mende,
K.Tóth,
E.Davioud-Charvet,
and
K.Becker
(2006).
Undressing of phosphine gold(I) complexes as irreversible inhibitors of human disulfide reductases.
|
| |
Angew Chem Int Ed Engl,
45,
1881-1886.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Nickel,
M.Trujillo,
S.Rahlfs,
M.Deponte,
R.Radi,
and
K.Becker
(2005).
Plasmodium falciparum 2-Cys peroxiredoxin reacts with plasmoredoxin and peroxynitrite.
|
| |
Biol Chem,
386,
1129-1136.
|
 |
|
|
|
|
 |
E.Cesareo,
L.J.Parker,
J.Z.Pedersen,
M.Nuccetelli,
A.P.Mazzetti,
A.Pastore,
G.Federici,
A.M.Caccuri,
G.Ricci,
J.J.Adams,
M.W.Parker,
and
M.Lo Bello
(2005).
Nitrosylation of human glutathione transferase P1-1 with dinitrosyl diglutathionyl iron complex in vitro and in vivo.
|
| |
J Biol Chem,
280,
42172-42180.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.I.Slobozhanina,
N.M.Kozlova,
L.M.Lukyanenko,
O.B.Oleksiuk,
R.Gabbianelli,
D.Fedeli,
G.C.Caulini,
and
G.Falcioni
(2005).
Lead-induced changes in human erythrocytes and lymphocytes.
|
| |
J Appl Toxicol,
25,
109-114.
|
 |
|
|
|
|
 |
R.G.Hu,
J.Sheng,
X.Qi,
Z.Xu,
T.T.Takahashi,
and
A.Varshavsky
(2005).
The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators.
|
| |
Nature,
437,
981-986.
|
 |
|
|
|
|
 |
A.Ledo,
J.Frade,
R.M.Barbosa,
and
J.Laranjinha
(2004).
Nitric oxide in brain: diffusion, targets and concentration dynamics in hippocampal subregions.
|
| |
Mol Aspects Med,
25,
75-89.
|
 |
|
|
|
|
 |
L.B.Poole,
P.A.Karplus,
and
A.Claiborne
(2004).
Protein sulfenic acids in redox signaling.
|
| |
Annu Rev Pharmacol Toxicol,
44,
325-347.
|
 |
|
|
|
|
 |
K.Becker,
S.Rahlfs,
C.Nickel,
and
R.H.Schirmer
(2003).
Glutathione--functions and metabolism in the malarial parasite Plasmodium falciparum.
|
| |
Biol Chem,
384,
551-566.
|
 |
|
|
|
|
 |
N.Campanale,
C.Nickel,
C.A.Daubenberger,
D.A.Wehlan,
J.J.Gorman,
N.Klonis,
K.Becker,
and
L.Tilley
(2003).
Identification and characterization of heme-interacting proteins in the malaria parasite, Plasmodium falciparum.
|
| |
J Biol Chem,
278,
27354-27361.
|
 |
|
|
|
|
 |
N.J.Costa,
C.C.Dahm,
F.Hurrell,
E.R.Taylor,
and
M.P.Murphy
(2003).
Interactions of mitochondrial thiols with nitric oxide.
|
| |
Antioxid Redox Signal,
5,
291-305.
|
 |
|
|
|
|
 |
C.A.Bottoms,
P.E.Smith,
and
J.J.Tanner
(2002).
A structurally conserved water molecule in Rossmann dinucleotide-binding domains.
|
| |
Protein Sci,
11,
2125-2137.
|
 |
|
|
|
|
 |
R.J.Mallis,
M.J.Hamann,
W.Zhao,
T.Zhang,
S.Hendrich,
and
J.A.Thomas
(2002).
Irreversible thiol oxidation in carbonic anhydrase III: protection by S-glutathiolation and detection in aging rats.
|
| |
Biol Chem,
383,
649-662.
|
 |
|
|
|
|
 |
S.O.Kim,
K.Merchant,
R.Nudelman,
W.F.Beyer,
T.Keng,
J.DeAngelo,
A.Hausladen,
and
J.S.Stamler
(2002).
OxyR: a molecular code for redox-related signaling.
|
| |
Cell,
109,
383-396.
|
 |
|
|
|
|
 |
A.Changela,
C.K.Ho,
A.Martins,
S.Shuman,
and
A.Mondragón
(2001).
Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme.
|
| |
EMBO J,
20,
2575-2586.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Fechner,
C.Böhme,
S.Gromer,
M.Funk,
R.Schirmer,
and
K.Becker
(2001).
Antioxidant status and nitric oxide in the malnutrition syndrome kwashiorkor.
|
| |
Pediatr Res,
49,
237-243.
|
 |
|
|
|
|
 |
D.Lando,
I.Pongratz,
L.Poellinger,
and
M.L.Whitelaw
(2000).
A redox mechanism controls differential DNA binding activities of hypoxia-inducible factor (HIF) 1alpha and the HIF-like factor.
|
| |
J Biol Chem,
275,
4618-4627.
|
 |
|
|
|
|
 |
E.Schröder,
J.A.Littlechild,
A.A.Lebedev,
N.Errington,
A.A.Vagin,
and
M.N.Isupov
(2000).
Crystal structure of decameric 2-Cys peroxiredoxin from human erythrocytes at 1.7 A resolution.
|
| |
Structure,
8,
605-615.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.P.Eu,
L.Liu,
M.Zeng,
and
J.S.Stamler
(2000).
An apoptotic model for nitrosative stress.
|
| |
Biochemistry,
39,
1040-1047.
|
 |
|
|
|
|
 |
J.Qin,
Y.Yang,
A.Velyvis,
and
A.Gronenborn
(2000).
Molecular views of redox regulation: three-dimensional structures of redox regulatory proteins and protein complexes.
|
| |
Antioxid Redox Signal,
2,
827-840.
|
 |
|
|
|
|
 |
K.Becker,
S.Gromer,
R.H.Schirmer,
and
S.Müller
(2000).
Thioredoxin reductase as a pathophysiological factor and drug target.
|
| |
Eur J Biochem,
267,
6118-6125.
|
 |
|
|
|
|
 |
K.V.Ramana,
B.L.Dixit,
S.Srivastava,
G.K.Balendiran,
S.K.Srivastava,
and
A.Bhatnagar
(2000).
Selective recognition of glutathiolated aldehydes by aldose reductase.
|
| |
Biochemistry,
39,
12172-12180.
|
 |
|
|
|
|
 |
P.Klatt,
and
S.Lamas
(2000).
Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress.
|
| |
Eur J Biochem,
267,
4928-4944.
|
 |
|
|
|
|
 |
T.Ueno,
and
T.Yoshimura
(2000).
The physiological activity and in vivo distribution of dinitrosyl dithiolato iron complex.
|
| |
Jpn J Pharmacol,
82,
95.
|
 |
|
|
|
|
 |
W.S.Cheung,
I.Bhan,
and
S.A.Lipton
(2000).
Nitric oxide (NO.) stabilizes whereas nitrosonium (NO+) enhances filopodial outgrowth by rat retinal ganglion cells in vitro.
|
| |
Brain Res,
868,
1.
|
 |
|
|
|
|
 |
B.Zech,
M.Wilm,
R.van Eldik,
and
B.Brüne
(1999).
Mass spectrometric analysis of nitric oxide-modified caspase-3.
|
| |
J Biol Chem,
274,
20931-20936.
|
 |
|
|
|
|
 |
P.Klatt,
E.P.Molina,
and
S.Lamas
(1999).
Nitric oxide inhibits c-Jun DNA binding by specifically targeted S-glutathionylation.
|
| |
J Biol Chem,
274,
15857-15864.
|
 |
|
|
|
|
 |
A.Hausladen,
A.J.Gow,
and
J.S.Stamler
(1998).
Nitrosative stress: metabolic pathway involving the flavohemoglobin.
|
| |
Proc Natl Acad Sci U S A,
95,
14100-14105.
|
 |
|
|
|
|
 |
J.S.Stamler,
and
A.Hausladen
(1998).
Oxidative modifications in nitrosative stress.
|
| |
Nat Struct Biol,
5,
247-249.
|
 |
|
|
|
|
 |
N.L.Chan,
P.H.Rogers,
and
A.Arnone
(1998).
Crystal structure of the S-nitroso form of liganded human hemoglobin.
|
| |
Biochemistry,
37,
16459-16464.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

