 |
PDBsum entry 1gln

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Aminoacyl-tRNA synthase
|
PDB id
|
|

|

|
1gln
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Aminoacyl-tRNA synthase
|
 |
|
Title:
|
 |
Architectures of class-defining and specific domains of glutamyl-tRNA synthetase
|
|
Structure:
|
 |
Glutamyl-tRNA synthetase. Chain: a
|
|
Source:
|
 |
Thermus thermophilus. Organism_taxid: 300852. Strain: hb8
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
O.Nureki,D.G.Vassylyev,K.Katayanagi,T.Shimizu,S.Sekine,T.Kigawa, T.Miyazawa,S.Yokoyama,K.Morikawa,Riken Structural Genomics/proteomics Initiative (Rsgi)
|
|
Key ref:
|
 |
O.Nureki
et al.
(1995).
Architectures of class-defining and specific domains of glutamyl-tRNA synthetase.
Science,
267,
1958-1965.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
20-Jul-94
|
Release date:
|
15-Oct-95
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P27000
(SYE_THET8) -
Glutamate--tRNA ligase from Thermus thermophilus (strain ATCC 27634 / DSM 579 / HB8)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
468 a.a.
468 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.17
- glutamate--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Glu) + L-glutamate + ATP = L-glutamyl-tRNA(Glu) + AMP + diphosphate
|
 |
 |
 |
 |
 |
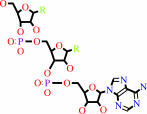 tRNA(Glu)
tRNA(Glu)
|
+
|
 L-glutamate
L-glutamate
|
+
|
 ATP
ATP
|
=
|
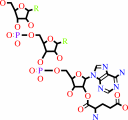 L-glutamyl-tRNA(Glu)
L-glutamyl-tRNA(Glu)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
267:1958-1965
(1995)
|
|
PubMed id:
|
|
|
|

|
| |
|
Architectures of class-defining and specific domains of glutamyl-tRNA synthetase.
|
|
O.Nureki,
D.G.Vassylyev,
K.Katayanagi,
T.Shimizu,
S.Sekine,
T.Kigawa,
T.Miyazawa,
S.Yokoyama,
K.Morikawa.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of a class I aminoacyl-transfer RNA synthetase,
glutamyl-tRNA synthetase (GluRS) from Thermus thermophilus, was solved and
refined at 2.5 A resolution. The amino-terminal half of GluRS shows a
geometrical similarity with that of Escherichia coli glutaminyl-tRNA synthetase
(GlnRS) of the same subclass in class I, comprising the class I-specific
Rossmann fold domain and the intervening subclass-specific alpha/beta domain.
These domains were found to have two GluRS-specific, secondary-structure
insertions, which then participated in the specific recognition of the D and
acceptor stems of tRNA(Glu) as indicated by mutagenesis analyses based on the
docking properties of GluRS and tRNA. In striking contrast to the beta-barrel
structure of the GlnRS carboxyl-terminal half, the GluRS carboxyl-terminal half
displayed an all-alpha-helix architecture, an alpha-helix cage, and mutagenesis
analyses indicated that it had a role in the anticodon recognition.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Kawai,
and
S.Yokoyama
(2010).
Professor Tatsuo Miyazawa: from molecular structure to biological function.
|
| |
J Biochem,
148,
631-638.
|
 |
|
|
|
|
 |
T.Ito,
N.Kiyasu,
R.Matsunaga,
S.Takahashi,
and
S.Yokoyama
(2010).
Structure of nondiscriminating glutamyl-tRNA synthetase from Thermotoga maritima.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
813-820.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.Fan,
and
J.S.Blanchard
(2009).
Toward the catalytic mechanism of a cysteine ligase (MshC) from Mycobacterium smegmatis: an enzyme involved in the biosynthetic pathway of mycothiol.
|
| |
Biochemistry,
48,
7150-7159.
|
 |
|
|
|
|
 |
T.T.Doan,
S.Natarajan,
H.Kim,
Y.J.Ahn,
J.G.Kim,
B.M.Lee,
and
L.W.Kang
(2009).
Cloning, expression, crystallization and preliminary X-ray crystallographic analysis of glutamyl-tRNA synthetase (Xoo1504) from Xanthomonas oryzae pv. oryzae.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
51-54.
|
 |
|
|
|
|
 |
L.W.Tremblay,
F.Fan,
M.W.Vetting,
and
J.S.Blanchard
(2008).
The 1.6 A crystal structure of Mycobacterium smegmatis MshC: the penultimate enzyme in the mycothiol biosynthetic pathway.
|
| |
Biochemistry,
47,
13326-13335.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Y.Dubois,
M.Blaise,
H.D.Becker,
V.Campanacci,
G.Keith,
R.Giegé,
C.Cambillau,
J.Lapointe,
and
D.Kern
(2004).
An aminoacyl-tRNA synthetase-like protein encoded by the Escherichia coli yadB gene glutamylates specifically tRNAAsp.
|
| |
Proc Natl Acad Sci U S A,
101,
7530-7535.
|
 |
|
|
|
|
 |
R.Banerjee,
D.Y.Dubois,
J.Gauthier,
S.X.Lin,
S.Roy,
and
J.Lapointe
(2004).
The zinc-binding site of a class I aminoacyl-tRNA synthetase is a SWIM domain that modulates amino acid binding via the tRNA acceptor arm.
|
| |
Eur J Biochem,
271,
724-733.
|
 |
|
|
|
|
 |
J.Cavarelli
(2003).
Pushing induced fit to its limits: tRNA-dependent active site assembly in class I aminoacyl-tRNA synthetases.
|
| |
Structure,
11,
484-486.
|
 |
|
|
|
|
 |
R.Banerjee,
A.K.Mandal,
R.Saha,
S.Guha,
S.Samaddar,
A.Bhattacharyya,
and
S.Roy
(2003).
Solvation change and ion release during aminoacylation by aminoacyl-tRNA synthetases.
|
| |
Nucleic Acids Res,
31,
6035-6042.
|
 |
|
|
|
|
 |
S.Sekine,
O.Nureki,
D.Y.Dubois,
S.Bernier,
R.Chênevert,
J.Lapointe,
D.G.Vassylyev,
and
S.Yokoyama
(2003).
ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding.
|
| |
EMBO J,
22,
676-688.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.J.Newberry,
Y.M.Hou,
and
J.J.Perona
(2002).
Structural origins of amino acid selection without editing by cysteinyl-tRNA synthetase.
|
| |
EMBO J,
21,
2778-2787.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Terada,
O.Nureki,
R.Ishitani,
A.Ambrogelly,
M.Ibba,
D.Söll,
and
S.Yokoyama
(2002).
Functional convergence of two lysyl-tRNA synthetases with unrelated topologies.
|
| |
Nat Struct Biol,
9,
257-262.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.von Delft,
A.Lewendon,
V.Dhanaraj,
T.L.Blundell,
C.Abell,
and
A.G.Smith
(2001).
The crystal structure of E. coli pantothenate synthetase confirms it as a member of the cytidylyltransferase superfamily.
|
| |
Structure,
9,
439-450.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Ribas de Pouplana,
and
P.Schimmel
(2001).
Two classes of tRNA synthetases suggested by sterically compatible dockings on tRNA acceptor stem.
|
| |
Cell,
104,
191-193.
|
 |
|
|
|
|
 |
O.Nureki,
S.Fukai,
S.Sekine,
A.Shimada,
T.Terada,
T.Nakama,
M.Shirouzu,
D.G.Vassylyev,
and
S.Yokoyama
(2001).
Structural basis for amino acid and tRNA recognition by class I aminoacyl-tRNA synthetases.
|
| |
Cold Spring Harb Symp Quant Biol,
66,
167-173.
|
 |
|
|
|
|
 |
C.R.Woese,
G.J.Olsen,
M.Ibba,
and
D.Söll
(2000).
Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process.
|
| |
Microbiol Mol Biol Rev,
64,
202-236.
|
 |
|
|
|
|
 |
E.Madore,
R.S.Lipman,
Y.M.Hou,
and
J.Lapointe
(2000).
Evidence for unfolding of the single-stranded GCCA 3'-End of a tRNA on its aminoacyl-tRNA synthetase from a stacked helical to a foldback conformation.
|
| |
Biochemistry,
39,
6791-6798.
|
 |
|
|
|
|
 |
I.Sugiura,
O.Nureki,
Y.Ugaji-Yoshikawa,
S.Kuwabara,
A.Shimada,
M.Tateno,
B.Lorber,
R.Giegé,
D.Moras,
S.Yokoyama,
and
M.Konno
(2000).
The 2.0 A crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules.
|
| |
Structure,
8,
197-208.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Ibba,
and
D.Soll
(2000).
Aminoacyl-tRNA synthesis.
|
| |
Annu Rev Biochem,
69,
617-650.
|
 |
|
|
|
|
 |
M.Kaminska,
M.Deniziak,
P.Kerjan,
J.Barciszewski,
and
M.Mirande
(2000).
A recurrent general RNA binding domain appended to plant methionyl-tRNA synthetase acts as a cis-acting cofactor for aminoacylation.
|
| |
EMBO J,
19,
6908-6917.
|
 |
|
|
|
|
 |
M.Praetorius-Ibba,
N.Stange-Thomann,
M.Kitabatake,
K.Ali,
I.Söll,
C.W.Carter,
M.Ibba,
and
D.Söll
(2000).
Ancient adaptation of the active site of tryptophanyl-tRNA synthetase for tryptophan binding.
|
| |
Biochemistry,
39,
13136-13143.
|
 |
|
|
|
|
 |
S.Onesti,
G.Desogus,
A.Brevet,
J.Chen,
P.Plateau,
S.Blanquet,
and
P.Brick
(2000).
Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding.
|
| |
Biochemistry,
39,
12853-12861.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.Guez,
S.Nair,
A.Chaffotte,
and
H.Bedouelle
(2000).
The anticodon-binding domain of tyrosyl-tRNA synthetase: state of folding and origin of the crystallographic disorder.
|
| |
Biochemistry,
39,
1739-1747.
|
 |
|
|
|
|
 |
C.H.Weber,
Y.S.Park,
S.Sanker,
C.Kent,
and
M.L.Ludwig
(1999).
A prototypical cytidylyltransferase: CTP:glycerol-3-phosphate cytidylyltransferase from bacillus subtilis.
|
| |
Structure,
7,
1113-1124.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.V.Franklund,
and
J.B.Goldberg
(1999).
Cloning of the glutamyl-tRNA synthetase (gltX) gene from Pseudomonas aeruginosa.
|
| |
J Bacteriol,
181,
3582-3586.
|
 |
|
|
|
|
 |
M.Sette,
R.Spurio,
P.van Tilborg,
C.O.Gualerzi,
and
R.Boelens
(1999).
Identification of the ribosome binding sites of translation initiation factor IF3 by multidimensional heteronuclear NMR spectroscopy.
|
| |
RNA,
5,
82-92.
|
 |
|
|
|
|
 |
S.Sekine,
O.Nureki,
M.Tateno,
and
S.Yokoyama
(1999).
The identity determinants required for the discrimination between tRNAGlu and tRNAAsp by glutamyl-tRNA synthetase from Escherichia coli.
|
| |
Eur J Biochem,
261,
354-360.
|
 |
|
|
|
|
 |
S.Watanabe,
T.Muramatsu,
H.Ao,
Y.Hirayama,
K.Takahashi,
M.Tanokura,
and
Y.Kuchino
(1999).
Molecular cloning of the Lon protease gene from Thermus thermophilus HB8 and characterization of its gene product.
|
| |
Eur J Biochem,
266,
811-819.
|
 |
|
|
|
|
 |
A.W.Curnow,
D.L.Tumbula,
J.T.Pelaschier,
B.Min,
and
D.Söll
(1998).
Glutamyl-tRNA(Gln) amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis.
|
| |
Proc Natl Acad Sci U S A,
95,
12838-12843.
|
 |
|
|
|
|
 |
C.Briand,
A.Poterszman,
A.Mitschler,
M.Yusupov,
J.C.Thierry,
and
D.Moras
(1998).
Crystals of Thermus thermophilus tRNAAsp complexed with its cognate aspartyl-tRNA synthetase have a solvent content of 75%. Comparison with other aminoacylation systems.
|
| |
Acta Crystallogr D Biol Crystallogr,
54,
1382-1386.
|
 |
|
|
|
|
 |
F.Agou,
S.Quevillon,
P.Kerjan,
and
M.Mirande
(1998).
Switching the amino acid specificity of an aminoacyl-tRNA synthetase.
|
| |
Biochemistry,
37,
11309-11314.
|
 |
|
|
|
|
 |
I.S.Day,
M.Golovkin,
and
A.S.Reddy
(1998).
Cloning of the cDNA for glutamyl-tRNA synthetase from Arabidopsis thaliana.
|
| |
Biochim Biophys Acta,
1399,
219-224.
|
 |
|
|
|
|
 |
J.Cavarelli,
B.Delagoutte,
G.Eriani,
J.Gangloff,
and
D.Moras
(1998).
L-arginine recognition by yeast arginyl-tRNA synthetase.
|
| |
EMBO J,
17,
5438-5448.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Ribas de Pouplana,
D.Buechter,
N.Y.Sardesai,
and
P.Schimmel
(1998).
Functional analysis of peptide motif for RNA microhelix binding suggests new family of RNA-binding domains.
|
| |
EMBO J,
17,
5449-5457.
|
 |
|
|
|
|
 |
R.S.Lipman,
and
Y.M.Hou
(1998).
Aminoacylation of tRNA in the evolution of an aminoacyl-tRNA synthetase.
|
| |
Proc Natl Acad Sci U S A,
95,
13495-13500.
|
 |
|
|
|
|
 |
V.L.Rath,
L.F.Silvian,
B.Beijer,
B.S.Sproat,
and
T.A.Steitz
(1998).
How glutaminyl-tRNA synthetase selects glutamine.
|
| |
Structure,
6,
439-449.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.H.McClain,
J.Schneider,
S.Bhattacharya,
and
K.Gabriel
(1998).
The importance of tRNA backbone-mediated interactions with synthetase for aminoacylation.
|
| |
Proc Natl Acad Sci U S A,
95,
460-465.
|
 |
|
|
|
|
 |
A.Aberg,
A.Yaremchuk,
M.Tukalo,
B.Rasmussen,
and
S.Cusack
(1997).
Crystal structure analysis of the activation of histidine by Thermus thermophilus histidyl-tRNA synthetase.
|
| |
Biochemistry,
36,
3084-3094.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.V.Efimov
(1997).
Structural trees for protein superfamilies.
|
| |
Proteins,
28,
241-260.
|
 |
|
|
|
|
 |
C.Cerini,
M.Semeriva,
and
D.Gratecos
(1997).
Evolution of the aminoacyl-tRNA synthetase family and the organization of the Drosophila glutamyl-prolyl-tRNA synthetase gene. Intron/exon structure of the gene, control of expression of the two mRNAs, selective advantage of the multienzyme complex.
|
| |
Eur J Biochem,
244,
176-185.
|
 |
|
|
|
|
 |
C.Chothia,
T.Hubbard,
S.Brenner,
H.Barns,
and
A.Murzin
(1997).
Protein folds in the all-beta and all-alpha classes.
|
| |
Annu Rev Biophys Biomol Struct,
26,
597-627.
|
 |
|
|
|
|
 |
E.F.Whelihan,
and
P.Schimmel
(1997).
Rescuing an essential enzyme-RNA complex with a non-essential appended domain.
|
| |
EMBO J,
16,
2968-2974.
|
 |
|
|
|
|
 |
E.Glasfeld,
and
P.Schimmel
(1997).
Zinc-dependent tRNA binding by a peptide element within a tRNA synthetase.
|
| |
Biochemistry,
36,
6739-6744.
|
 |
|
|
|
|
 |
E.Liepinsh,
M.Andersson,
J.M.Ruysschaert,
and
G.Otting
(1997).
Saposin fold revealed by the NMR structure of NK-lysin.
|
| |
Nat Struct Biol,
4,
793-795.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Zhou,
E.D.Wang,
R.L.Campbell,
Y.L.Wang,
and
S.X.Lin
(1997).
Crystallization and preliminary X-ray diffraction analysis of arginyl-tRNA synthetase from Escherichia coli.
|
| |
Protein Sci,
6,
2636-2638.
|
 |
|
|
|
|
 |
R.Kato,
K.Hasegawa,
Y.Hidaka,
S.Kuramitsu,
and
T.Hoshino
(1997).
Characterization of a thermostable DNA photolyase from an extremely thermophilic bacterium, Thermus thermophilus HB27.
|
| |
J Bacteriol,
179,
6499-6503.
|
 |
|
|
|
|
 |
E.Glasfeld,
J.A.Landro,
and
P.Schimmel
(1996).
C-terminal zinc-containing peptide required for RNA recognition by a class I tRNA synthetase.
|
| |
Biochemistry,
35,
4139-4145.
|
 |
|
|
|
|
 |
J.A.Moore,
A.Chen,
M.Yan,
A.P.Hurlburt,
and
C.D.Poulter
(1996).
Identification of the gltX gene encoding glutamyl-tRNA synthetase from Methanobacterium thermoautotrophicum.
|
| |
Biochim Biophys Acta,
1305,
113-116.
|
 |
|
|
|
|
 |
L.Lin,
and
P.Schimmel
(1996).
Mutational analysis suggests the same design for editing activities of two tRNA synthetases.
|
| |
Biochemistry,
35,
5596-5601.
|
 |
|
|
|
|
 |
M.H.Mazauric,
J.Reinbolt,
B.Lorber,
C.Ebel,
G.Keith,
R.Giegé,
and
D.Kern
(1996).
An example of non-conservation of oligomeric structure in prokaryotic aminoacyl-tRNA synthetases. Biochemical and structural properties of glycyl-tRNA synthetase from Thermus thermophilus.
|
| |
Eur J Biochem,
241,
814-826.
|
 |
|
|
|
|
 |
M.Sassanfar,
J.E.Kranz,
P.Gallant,
P.Schimmel,
and
K.Shiba
(1996).
A eubacterial Mycobacterium tuberculosis tRNA synthetase is eukaryote-like and resistant to a eubacterial-specific antisynthetase drug.
|
| |
Biochemistry,
35,
9995.
|
 |
|
|
|
|
 |
S.P.Hale,
and
P.Schimmel
(1996).
Protein synthesis editing by a DNA aptamer.
|
| |
Proc Natl Acad Sci U S A,
93,
2755-2758.
|
 |
|
|
|
|
 |
S.Sever,
K.Rogers,
M.J.Rogers,
C.Carter,
and
D.Söll
(1996).
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer interface in optimizing amino acid binding.
|
| |
Biochemistry,
35,
32-40.
|
 |
|
|
|
|
 |
E.Schmitt,
M.Panvert,
S.Blanquet,
and
Y.Mechulam
(1995).
Transition state stabilization by the 'high' motif of class I aminoacyl-tRNA synthetases: the case of Escherichia coli methionyl-tRNA synthetase.
|
| |
Nucleic Acids Res,
23,
4793-4798.
|
 |
|
|
|
|
 |
J.Guevara,
E.T.Walch,
H.F.Epstein,
J.T.Sparrow,
A.M.Gotto,
and
N.V.Valentinova
(1995).
Evidence that apoB-100 of low-density lipoproteins is a novel Src-related protein kinase.
|
| |
J Protein Chem,
14,
627-631.
|
 |
|
|
|
|
 |
P.Schimmel,
and
L.Ribas de Pouplana
(1995).
Transfer RNA: from minihelix to genetic code.
|
| |
Cell,
81,
983-986.
|
 |
|
|
|
|
 |
S.Cusack
(1995).
Eleven down and nine to go.
|
| |
Nat Struct Biol,
2,
824-831.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

