
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 511 a.a.
511 a.a.
|
 |

|

|

|

|

|

|

|

 388 a.a.
388 a.a.
|
 |

|

|

|

|

|

|

|

 166 a.a.
166 a.a.
|
 |

|

|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Methane monooxygenase hydroxylase, form ii crystallized anaerobically from reduced enzyme
|
|
Structure:
|
 |
Methane monooxygenase component a, alpha chain. Chain: a, b. Synonym: hydroxylase alpha subunit. Methane monooxygenase component a, beta chain. Chain: c, d. Synonym: hydroxylase beta subunit, methane monooxygenase a beta chain. Methane monooxygenase component a, gamma chain. Chain: e, f.
|
|
Source:
|
 |
Methylococcus capsulatus. Organism_taxid: 414. Organism_taxid: 414
|
|
Biol. unit:
|
 |
Hexamer (from
)
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.218
|
R-free:
|
0.253
|
|
|
Authors:
|
 |
D.A.Whittington,S.J.Lippard
|
|
Key ref:
|
 |
D.A.Whittington
and
S.J.Lippard
(2001).
Crystal structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath) demonstrating geometrical variability at the dinuclear iron active site.
J Am Chem Soc,
123,
827-838.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
03-Oct-00
|
Release date:
|
28-Feb-01
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P22869
(MEMA_METCA) -
Methane monooxygenase component A alpha chain from Methylococcus capsulatus (strain ATCC 33009 / NCIMB 11132 / Bath)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
527 a.a.
511 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F:
E.C.1.14.13.25
- methane monooxygenase (soluble).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
methane + NADH + O2 + H+ = methanol + NAD+ + H2O
|
|
2.
|
methane + NADPH + O2 + H+ = methanol + NADP+ + H2O
|
|
 |
 |
 |
 |
 |
methane
|
+
|
 NADH
NADH
|
+
|
 O2
O2
|
+
|
H(+)
|
=
|
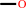 methanol
methanol
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
methane
|
+
|
 NADPH
NADPH
|
+
|
 O2
O2
|
+
|
H(+)
|
=
|
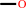 methanol
methanol
|
+
|
 NADP(+)
NADP(+)
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Am Chem Soc
123:827-838
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath) demonstrating geometrical variability at the dinuclear iron active site.
|
|
D.A.Whittington,
S.J.Lippard.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The oxidation of methane to methanol is performed at carboxylate-bridged
dinuclear iron centers in the soluble methane monooxygenase hydroxylase (MMOH).
Previous structural studies of MMOH, and the related R2 subunit of
ribonucleotide reductase, have demonstrated the occurrence of carboxylate shifts
involving glutamate residues that ligate the catalytic iron atoms. These shifts
are thought to have important mechanistic implications. Recent kinetic and
theoretical studies have also emphasized the importance of hydrogen bonding and
pH effects at the active site. We report here crystal structures of MMOH from
Methylococcus capsulatus (Bath) in the diiron(II), diiron(III), and mixed-valent
Fe(II)Fe(III) oxidation states, and at pH values of 6.2, 7.0, and 8.5. These
structures were investigated in an effort to delineate the range of possible
motions at the MMOH active site and to identify hydrogen-bonding interactions
that may be important in understanding catalysis by the enzyme. Our results
present the first view of the diiron center in the mixed-valent state, and they
indicate an increased lability for ferrous ions in the enzyme. Alternate
conformations of Asn214 near the active site according to redox state and a
distortion in one of the alpha-helices adjacent to the metal center in the
diiron(II) state have also been identified. These changes alter the surface of
the protein in the vicinity of the catalytic core and may have implications for
small-molecule accessibility to the active site and for protein component
interactions in the methane monooxygenase system. Collectively, these results
help to explain previous spectroscopic observations and provide new insight into
catalysis by the enzyme.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.J.Lee,
M.S.McCormick,
S.J.Lippard,
and
U.S.Cho
(2013).
Control of substrate access to the active site in methane monooxygenase.
|
| |
Nature,
494,
380-384.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.B.Bell,
J.R.Calhoun,
E.Bobyr,
P.P.Wei,
B.Hedman,
K.O.Hodgson,
W.F.Degrado,
and
E.I.Solomon
(2009).
Spectroscopic definition of the biferrous and biferric sites in de novo designed four-helix bundle DFsc peptides: implications for O2 reactivity of binuclear non-heme iron enzymes.
|
| |
Biochemistry,
48,
59-73.
|
 |
|
|
|
|
 |
C.E.Tinberg,
and
S.J.Lippard
(2009).
Revisiting the mechanism of dioxygen activation in soluble methane monooxygenase from M. capsulatus (Bath): evidence for a multi-step, proton-dependent reaction pathway.
|
| |
Biochemistry,
48,
12145-12158.
|
 |
|
|
|
|
 |
I.Siewert,
and
C.Limberg
(2009).
Low-molecular-weight analogues of the soluble methane monooxygenase (sMMO): from the structural mimicking of resting states and intermediates to functional models.
|
| |
Chemistry,
15,
10316-10328.
|
 |
|
|
|
|
 |
J.R.Frisch,
V.V.Vu,
M.Martinho,
E.Münck,
and
L.Que
(2009).
Characterization of two distinct adducts in the reaction of a nonheme diiron(II) complex with O2.
|
| |
Inorg Chem,
48,
8325-8336.
|
 |
|
|
|
|
 |
J.K.Schwartz,
X.S.Liu,
T.Tosha,
E.C.Theil,
and
E.I.Solomon
(2008).
Spectroscopic definition of the ferroxidase site in M ferritin: comparison of binuclear substrate vs cofactor active sites.
|
| |
J Am Chem Soc,
130,
9441-9450.
|
 |
|
|
|
|
 |
N.Mitić,
J.K.Schwartz,
B.J.Brazeau,
J.D.Lipscomb,
and
E.I.Solomon
(2008).
CD and MCD studies of the effects of component B variant binding on the biferrous active site of methane monooxygenase.
|
| |
Biochemistry,
47,
8386-8397.
|
 |
|
|
|
|
 |
D.Rinaldo,
D.M.Philipp,
S.J.Lippard,
and
R.A.Friesner
(2007).
Intermediates in dioxygen activation by methane monooxygenase: a QM/MM study.
|
| |
J Am Chem Soc,
129,
3135-3147.
|
 |
|
|
|
|
 |
J.E.Guy,
E.Whittle,
D.Kumaran,
Y.Lindqvist,
and
J.Shanklin
(2007).
The crystal structure of the ivy Delta4-16:0-ACP desaturase reveals structural details of the oxidized active site and potential determinants of regioselectivity.
|
| |
J Biol Chem,
282,
19863-19871.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.S.Nesterov,
V.N.Kokozay,
V.V.Dyakonenko,
O.V.Shishkin,
J.Jezierska,
A.Ozarowski,
A.M.Kirillov,
M.N.Kopylovich,
and
A.J.Pombeiro
(2006).
An unprecedented heterotrimetallic Fe/Cu/Co core for mild and highly efficient catalytic oxidation of cycloalkanes by hydrogen peroxide.
|
| |
Chem Commun (Camb),
(),
4605-4607.
|
 |
|
|
|
|
 |
E.C.Carson,
and
S.J.Lippard
(2006).
Synthesis, characterization, and preliminary oxygenation studies of benzyl- and ethyl-substituted pyridine ligands of carboxylate-rich diiron(II) complexes.
|
| |
Inorg Chem,
45,
828-836.
|
 |
|
|
|
|
 |
E.C.Carson,
and
S.J.Lippard
(2006).
Dioxygen-initiated oxidation of heteroatomic substrates incorporated into ancillary pyridine ligands of carboxylate-rich diiron(II) complexes.
|
| |
Inorg Chem,
45,
837-848.
|
 |
|
|
|
|
 |
K.H.Halsey,
L.A.Sayavedra-Soto,
P.J.Bottomley,
and
D.J.Arp
(2006).
Site-directed amino acid substitutions in the hydroxylase alpha subunit of butane monooxygenase from Pseudomonas butanovora: Implications for substrates knocking at the gate.
|
| |
J Bacteriol,
188,
4962-4969.
|
 |
|
|
|
|
 |
M.H.Sazinsky,
P.W.Dunten,
M.S.McCormick,
A.DiDonato,
and
S.J.Lippard
(2006).
X-ray structure of a hydroxylase-regulatory protein complex from a hydrocarbon-oxidizing multicomponent monooxygenase, Pseudomonas sp. OX1 phenol hydroxylase.
|
| |
Biochemistry,
45,
15392-15404.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.S.McCormick,
M.H.Sazinsky,
K.L.Condon,
and
S.J.Lippard
(2006).
X-ray crystal structures of manganese(II)-reconstituted and native toluene/o-xylene monooxygenase hydroxylase reveal rotamer shifts in conserved residues and an enhanced view of the protein interior.
|
| |
J Am Chem Soc,
128,
15108-15110.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.M.Brown,
T.T.Caradoc-Davies,
J.M.Dickson,
G.J.Cooper,
K.M.Loomes,
and
E.N.Baker
(2006).
Crystal structure of a substrate complex of myo-inositol oxygenase, a di-iron oxygenase with a key role in inositol metabolism.
|
| |
Proc Natl Acad Sci U S A,
103,
15032-15037.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Yoon,
and
S.J.Lippard
(2006).
Mechanistic studies of the oxidative N-dealkylation of a substrate tethered to carboxylate-bridged diiron(II) complexes, [Fe2(mu-O2CAr(Tol))2(O2CAr(Tol))2(N,N-Bn2en)2].
|
| |
Inorg Chem,
45,
5438-5446.
|
 |
|
|
|
|
 |
J.L.Blazyk,
G.T.Gassner,
and
S.J.Lippard
(2005).
Intermolecular electron-transfer reactions in soluble methane monooxygenase: a role for hysteresis in protein function.
|
| |
J Am Chem Soc,
127,
17364-17376.
|
 |
|
|
|
|
 |
J.R.Calhoun,
F.Nastri,
O.Maglio,
V.Pavone,
A.Lombardi,
and
W.F.DeGrado
(2005).
Artificial diiron proteins: from structure to function.
|
| |
Biopolymers,
80,
264-278.
|
 |
|
|
|
|
 |
R.A.Friesner,
and
V.Guallar
(2005).
Ab initio quantum chemical and mixed quantum mechanics/molecular mechanics (QM/MM) methods for studying enzymatic catalysis.
|
| |
Annu Rev Phys Chem,
56,
389-427.
|
 |
|
|
|
|
 |
K.R.Strand,
S.Karlsen,
M.Kolberg,
A.K.Røhr,
C.H.Görbitz,
and
K.K.Andersson
(2004).
Crystal structural studies of changes in the native dinuclear iron center of ribonucleotide reductase protein R2 from mouse.
|
| |
J Biol Chem,
279,
46794-46801.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Ward,
Ã.˜.Larsen,
J.Sakwa,
L.Bruseth,
H.Khouri,
A.S.Durkin,
G.Dimitrov,
L.Jiang,
D.Scanlan,
K.H.Kang,
M.Lewis,
K.E.Nelson,
B.Methé,
M.Wu,
J.F.Heidelberg,
I.T.Paulsen,
D.Fouts,
J.Ravel,
H.Tettelin,
Q.Ren,
T.Read,
R.T.DeBoy,
R.Seshadri,
S.L.Salzberg,
H.B.Jensen,
N.K.Birkeland,
W.C.Nelson,
R.J.Dodson,
S.H.Grindhaug,
I.Holt,
I.Eidhammer,
I.Jonasen,
S.Vanaken,
T.Utterback,
T.V.Feldblyum,
C.M.Fraser,
J.R.Lillehaug,
and
J.A.Eisen
(2004).
Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath).
|
| |
PLoS Biol,
2,
e303.
|
 |
|
|
|
|
 |
W.C.Kao,
Y.R.Chen,
E.C.Yi,
H.Lee,
Q.Tian,
K.M.Wu,
S.F.Tsai,
S.S.Yu,
Y.J.Chen,
R.Aebersold,
and
S.I.Chan
(2004).
Quantitative proteomic analysis of metabolic regulation by copper ions in Methylococcus capsulatus (Bath).
|
| |
J Biol Chem,
279,
51554-51560.
|
 |
|
|
|
|
 |
A.Glasfeld,
E.Guedon,
J.D.Helmann,
and
R.G.Brennan
(2003).
Structure of the manganese-bound manganese transport regulator of Bacillus subtilis.
|
| |
Nat Struct Biol,
10,
652-657.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.A.Kopp,
E.A.Berg,
C.E.Costello,
and
S.J.Lippard
(2003).
Structural features of covalently cross-linked hydroxylase and reductase proteins of soluble methane monooxygenase as revealed by mass spectrometric analysis.
|
| |
J Biol Chem,
278,
20939-20945.
|
 |
|
|
|
|
 |
M.Moche,
J.Shanklin,
A.Ghoshal,
and
Y.Lindqvist
(2003).
Azide and acetate complexes plus two iron-depleted crystal structures of the di-iron enzyme delta9 stearoyl-acyl carrier protein desaturase. Implications for oxygen activation and catalytic intermediates.
|
| |
J Biol Chem,
278,
25072-25080.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.Maglio,
F.Nastri,
V.Pavone,
A.Lombardi,
and
W.F.DeGrado
(2003).
Preorganization of molecular binding sites in designed diiron proteins.
|
| |
Proc Natl Acad Sci U S A,
100,
3772-3777.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.A.Kopp,
and
S.J.Lippard
(2002).
Soluble methane monooxygenase: activation of dioxygen and methane.
|
| |
Curr Opin Chem Biol,
6,
568-576.
|
 |
|
|
|
|
 |
K.R.Strand,
S.Karlsen,
and
K.K.Andersson
(2002).
Cobalt substitution of mouse R2 ribonucleotide reductase as a model for the reactive diferrous state Spectroscopic and structural evidence for a ferromagnetically coupled dinuclear cobalt cluster.
|
| |
J Biol Chem,
277,
34229-34238.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Que,
and
W.B.Tolman
(2002).
Bis(mu-oxo)dimetal "diamond" cores in copper and iron complexes relevant to biocatalysis.
|
| |
Angew Chem Int Ed Engl,
41,
1114-1137.
|
 |
|
|
|
|
 |
M.J.Ryle,
and
R.P.Hausinger
(2002).
Non-heme iron oxygenases.
|
| |
Curr Opin Chem Biol,
6,
193-201.
|
 |
|
|
|
|
 |
M.Merkx,
and
S.J.Lippard
(2002).
Why OrfY? Characterization of MMOD, a long overlooked component of the soluble methane monooxygenase from Methylococcus capsulatus (Bath).
|
| |
J Biol Chem,
277,
5858-5865.
|
 |
|
|
|
|
 |
V.Guallar,
B.F.Gherman,
S.J.Lippard,
and
R.A.Friesner
(2002).
Quantum chemical studies of methane monooxygenase: comparision with P450.
|
| |
Curr Opin Chem Biol,
6,
236-242.
|
 |
|
|
|
|
 |
M.Merkx,
D.A.Kopp,
M.H.Sazinsky,
J.L.Blazyk,
J.Müller,
and
S.J.Lippard
(2001).
Dioxygen Activation and Methane Hydroxylation by Soluble Methane Monooxygenase: A Tale of Two Irons and Three Proteins A list of abbreviations can be found in Section 7.
|
| |
Angew Chem Int Ed Engl,
40,
2782-2807.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |
|

