
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.1.14.11.28
- proline 3-hydroxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-proline + 2-oxoglutarate + O2 = cis-3-hydroxy-L-proline + succinate + CO2
|
 |
 |
 |
 |
 |
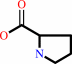 L-proline
L-proline
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
+
|
 O2
O2
|
=
|
cis-3-hydroxy-L-proline
|
+
|
 succinate
succinate
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Eur J Biochem
268:6625-6636
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of proline 3-hydroxylase. Evolution of the family of 2-oxoglutarate dependent oxygenases.
|
|
I.J.Clifton,
L.C.Hsueh,
J.E.Baldwin,
K.Harlos,
C.J.Schofield.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Iron (II)/2-oxoglutarate (2-OG)-dependent oxygenases catalyse oxidative
reactions in a range of metabolic processes including the hydroxylation of
proline and lysine residues during the post-translational modification of
collagen. 2-OG oxygenases commonly require ascorbate for full activity. In the
vitamin C deficient disease, scurvy, reduced activity of 2-OG oxygenases results
in impaired formation of collagen. Here we report the crystal structure of
bacterial proline 3-hydroxylase from Streptomyces sp., an enzyme which
hydroxylates proline at position 3, the first of a 2-OG oxygenase catalysing
oxidation of a free alpha-amino acid. Structures were obtained for the enzyme in
the absence of iron (to 2.3A resolution, R=20.2%, Rfree=25.3%) and that
complexed to iron (II) (to 2.4A resolution, R=19.8%, Rfree=22.6%). The structure
contains conserved motifs present in other 2-OG oxygenases including a 'jelly
roll' beta strand core and residues binding iron and 2-oxoglutarate, consistent
with divergent evolution within the extended family. The structure differs
significantly from many other 2-OG oxygenases in possessing a discrete
C-terminal helical domain. Analysis of the structure suggests a model for
proline binding and a mechanism for uncoupling of proline and 2-OG turnover.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1 Reaction summary. Reactions catalysed by
mononuclear nonhaem iron oxygenases (A) proline 3-hydroxylase
(P-3-H) (B) proline-4-hydroxylase (C) one of the reactions
catalysed by deacetoxycephalosporin C synthase (DAOCS) (D)
isopenicillin N synthase (IPNS) (E) clavaminic acid synthase
(CAS) (F) p-hydroxyphenylpyruvate dioxygenase (HPPD).
|
 |
Figure 4.
Fig. 4 View of the P-3-H active site. The active site of
the enzyme (molecule A) cocrystallized with iron(II) sulfate.
The  -barrel core
is in green, the iron binding ligands in purple, the iron in
orange and the N-terminal region and C-terminal domain in
magenta. The ferrous iron is ligated by the side chains of
His107, Asp109 and His158. Note: (a) Arg168, Ser170 and His135
which are probably involved in binding 2-OG; (b) Arg95, Arg97,
Arg122 and His43 which may bind the proline carboxylate; (c) the
disordered loop (in orange) containing six sequential acidic
residues, which may bind the imino group of proline. -barrel core
is in green, the iron binding ligands in purple, the iron in
orange and the N-terminal region and C-terminal domain in
magenta. The ferrous iron is ligated by the side chains of
His107, Asp109 and His158. Note: (a) Arg168, Ser170 and His135
which are probably involved in binding 2-OG; (b) Arg95, Arg97,
Arg122 and His43 which may bind the proline carboxylate; (c) the
disordered loop (in orange) containing six sequential acidic
residues, which may bind the imino group of proline.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Federation of European Biochemical Societies:
Eur J Biochem
(2001,
268,
6625-6636)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.L.Gorres,
and
R.T.Raines
(2010).
Prolyl 4-hydroxylase.
|
| |
Crit Rev Biochem Mol Biol,
45,
106-124.
|
 |
|
|
|
|
 |
L.M.Blank,
B.E.Ebert,
K.Buehler,
and
B.Bühler
(2010).
Redox biocatalysis and metabolism: molecular mechanisms and metabolic network analysis.
|
| |
Antioxid Redox Signal,
13,
349-394.
|
 |
|
|
|
|
 |
M.A.Culpepper,
E.E.Scott,
and
J.Limburg
(2010).
Crystal structure of prolyl 4-hydroxylase from Bacillus anthracis.
|
| |
Biochemistry,
49,
124-133.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.M.Iyer,
M.Tahiliani,
A.Rao,
and
L.Aravind
(2009).
Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids.
|
| |
Cell Cycle,
8,
1698-1710.
|
 |
|
|
|
|
 |
M.P.Dunn,
and
A.Di Gregorio
(2009).
The evolutionarily conserved leprecan gene: its regulation by Brachyury and its role in the developing Ciona notochord.
|
| |
Dev Biol,
328,
561-574.
|
 |
|
|
|
|
 |
R.Chowdhury,
M.A.McDonough,
J.Mecinović,
C.Loenarz,
E.Flashman,
K.S.Hewitson,
C.Domene,
and
C.J.Schofield
(2009).
Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases.
|
| |
Structure,
17,
981-989.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Lohkamp,
and
D.Dobritzsch
(2008).
A mixture of fortunes: the curious determination of the structure of Escherichia coli BL21 Gab protein.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
407-415.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.G.Kovaleva,
and
J.D.Lipscomb
(2008).
Versatility of biological non-heme Fe(II) centers in oxygen activation reactions.
|
| |
Nat Chem Biol,
4,
186-193.
|
 |
|
|
|
|
 |
M.K.Koski,
R.Hieta,
C.Böllner,
K.I.Kivirikko,
J.Myllyharju,
and
R.K.Wierenga
(2007).
The active site of an algal prolyl 4-hydroxylase has a large structural plasticity.
|
| |
J Biol Chem,
282,
37112-37123.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.Purpero,
and
G.R.Moran
(2007).
The diverse and pervasive chemistries of the alpha-keto acid dependent enzymes.
|
| |
J Biol Inorg Chem,
12,
587-601.
|
 |
|
|
|
|
 |
A.Daruzzaman,
I.J.Clifton,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2006).
Unexpected oxidation of a depsipeptide substrate analogue in crystalline isopenicillin N synthase.
|
| |
Chembiochem,
7,
351-358.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.D.Koehntop,
S.Marimanikkuppam,
M.J.Ryle,
R.P.Hausinger,
and
L.Que
(2006).
Self-hydroxylation of taurine/alpha-ketoglutarate dioxygenase: evidence for more than one oxygen activation mechanism.
|
| |
J Biol Inorg Chem,
11,
63-72.
|
 |
|
|
|
|
 |
M.A.McDonough,
V.Li,
E.Flashman,
R.Chowdhury,
C.Mohr,
B.M.Liénard,
J.Zondlo,
N.J.Oldham,
I.J.Clifton,
J.Lewis,
L.A.McNeill,
R.J.Kurzeja,
K.S.Hewitson,
E.Yang,
S.Jordan,
R.S.Syed,
and
C.J.Schofield
(2006).
Cellular oxygen sensing: Crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2).
|
| |
Proc Natl Acad Sci U S A,
103,
9814-9819.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.D.Guzy,
and
P.T.Schumacker
(2006).
Oxygen sensing by mitochondria at complex III: the paradox of increased reactive oxygen species during hypoxia.
|
| |
Exp Physiol,
91,
807-819.
|
 |
|
|
|
|
 |
T.A.Müller,
M.I.Zavodszky,
M.Feig,
L.A.Kuhn,
and
R.P.Hausinger
(2006).
Structural basis for the enantiospecificities of R- and S-specific phenoxypropionate/alpha-ketoglutarate dioxygenases.
|
| |
Protein Sci,
15,
1356-1368.
|
 |
|
|
|
|
 |
L.A.McNeill,
E.Flashman,
M.R.Buck,
K.S.Hewitson,
I.J.Clifton,
G.Jeschke,
T.D.Claridge,
D.Ehrismann,
N.J.Oldham,
and
C.J.Schofield
(2005).
Hypoxia-inducible factor prolyl hydroxylase 2 has a high affinity for ferrous iron and 2-oxoglutarate.
|
| |
Mol Biosyst,
1,
321-324.
|
 |
|
|
|
|
 |
M.A.McDonough,
K.L.Kavanagh,
D.Butler,
T.Searls,
U.Oppermann,
and
C.J.Schofield
(2005).
Structure of human phytanoyl-CoA 2-hydroxylase identifies molecular mechanisms of Refsum disease.
|
| |
J Biol Chem,
280,
41101-41110.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Hansen,
B.Schlichting,
M.Felgendreher,
and
P.Schönheit
(2005).
Cupin-type phosphoglucose isomerases (Cupin-PGIs) constitute a novel metal-dependent PGI family representing a convergent line of PGI evolution.
|
| |
J Bacteriol,
187,
1621-1631.
|
 |
|
|
|
|
 |
J.S.Hsu,
Y.B.Yang,
C.H.Deng,
C.L.Wei,
S.H.Liaw,
and
Y.C.Tsai
(2004).
Family shuffling of expandase genes to enhance substrate specificity for penicillin G.
|
| |
Appl Environ Microbiol,
70,
6257-6263.
|
 |
|
|
|
|
 |
K.Valegård,
A.C.Terwisscha van Scheltinga,
A.Dubus,
G.Ranghino,
L.M.Oster,
J.Hajdu,
and
I.Andersson
(2004).
The structural basis of cephalosporin formation in a mononuclear ferrous enzyme.
|
| |
Nat Struct Mol Biol,
11,
95.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Lee,
S.J.Kim,
D.G.Jeong,
S.M.Lee,
and
S.E.Ryu
(2003).
Structure of human FIH-1 reveals a unique active site pocket and interaction sites for HIF-1 and von Hippel-Lindau.
|
| |
J Biol Chem,
278,
7558-7563.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.J.Clifton,
L.X.Doan,
M.C.Sleeman,
M.Topf,
H.Suzuki,
R.C.Wilmouth,
and
C.J.Schofield
(2003).
Crystal structure of carbapenem synthase (CarC).
|
| |
J Biol Chem,
278,
20843-20850.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Mukherji,
C.J.Schofield,
A.S.Wierzbicki,
G.A.Jansen,
R.J.Wanders,
and
M.D.Lloyd
(2003).
The chemical biology of branched-chain lipid metabolism.
|
| |
Prog Lipid Res,
42,
359-376.
|
 |
|
|
|
|
 |
C.E.Dann,
R.K.Bruick,
and
J.Deisenhofer
(2002).
Structure of factor-inhibiting hypoxia-inducible factor 1: An asparaginyl hydroxylase involved in the hypoxic response pathway.
|
| |
Proc Natl Acad Sci U S A,
99,
15351-15356.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Lando,
D.J.Peet,
J.J.Gorman,
D.A.Whelan,
M.L.Whitelaw,
and
R.K.Bruick
(2002).
FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor.
|
| |
Genes Dev,
16,
1466-1471.
|
 |
|
|
|
|
 |
J.C.Dunning Hotopp,
and
R.P.Hausinger
(2002).
Probing the 2,4-dichlorophenoxyacetate/alpha-ketoglutarate dioxygenase substrate-binding site by site-directed mutagenesis and mechanism-based inactivation.
|
| |
Biochemistry,
41,
9787-9794.
|
 |
|
|
|
|
 |
J.H.Min,
H.Yang,
M.Ivan,
F.Gertler,
W.G.Kaelin,
and
N.P.Pavletich
(2002).
Structure of an HIF-1alpha -pVHL complex: hydroxyproline recognition in signaling.
|
| |
Science,
296,
1886-1889.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Ryle,
and
R.P.Hausinger
(2002).
Non-heme iron oxygenases.
|
| |
Curr Opin Chem Biol,
6,
193-201.
|
 |
|
|
|
|
 |
R.Anand,
P.C.Dorrestein,
C.Kinsland,
T.P.Begley,
and
S.E.Ealick
(2002).
Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution.
|
| |
Biochemistry,
41,
7659-7669.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

