 |
PDBsum entry 1dm3

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.9
- acetyl-CoA C-acetyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Mevalonate Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 acetyl-CoA = acetoacetyl-CoA + CoA
|
 |
 |
 |
 |
 |
2
×
acetyl-CoA
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
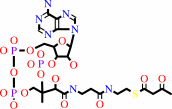 acetoacetyl-CoA
acetoacetyl-CoA
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
297:1171-1182
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic analysis of the reaction pathway of Zoogloea ramigera biosynthetic thiolase.
|
|
Y.Modis,
R.K.Wierenga.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Biosynthetic thiolases catalyze the biological Claisen condensation of two
acetyl-CoA molecules to form acetoacetyl-CoA. This is one of the fundamental
categories of carbon skeletal assembly patterns in biological systems and is the
first step in many biosynthetic pathways including those which generate
cholesterol, steroid hormones and ketone body energy storage molecules. High
resolution crystal structures of the tetrameric biosynthetic thiolase from
Zoogloea ramigera were determined (i) in the absence of active site ligands,
(ii) in the presence of CoA, and (iii) from protein crystals which were flash
frozen after a short soak with acetyl-CoA, the enzyme's substrate in the
biosynthetic reaction. In the latter structure, a reaction intermediate was
trapped: the enzyme was found to be acetylated at Cys89 and a molecule of
acetyl-CoA was bound in the active site pocket. A comparison of the three new
structures and the two previously published thiolase structures reveals that
small adjustments in the conformation of the acetylated Cys89 side-chain allow
CoA and acetyl-CoA to adopt identical modes of binding. The proximity of the
acetyl moiety of acetyl-CoA to the sulfur atom of Cys378 supports the hypothesis
that Cys378 is important for proton exchange in both steps of the reaction. The
thioester oxygen atom of the acetylated enzyme points into an oxyanion hole
formed by the nitrogen atoms of Cys89 and Gly380, thus facilitating the
condensation reaction. The interaction between the thioester oxygen atom of
acetyl-CoA and His348 assists the condensation step of catalysis by stabilizing
a negative charge on the thioester oxygen atom. Our structure of acetyl-CoA
bound to thiolase also highlights the importance in catalysis of a hydrogen
bonding network between Cys89 and Cys378, which includes the thioester oxygen
atom of acetyl-CoA, and extends from the catalytic site through the enzyme to
the opposite molecular surface. This hydrogen bonding network is different in
yeast degradative thiolase, indicating that the catalytic properties of each
enzyme may be modulated by differences in their hydrogen bonding networks.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Stereo picture of the biosynthetic thiolase
tetramer with its "tetrahedral cage" tetramerization motif. The
tetramer is colored according to the B-factor of the C^a atom of
each residue. Dark blue corresponds to a B-factor of  vert,
similar 5 Å2; red corresponds to 100 Å2. The A-B
dimer (upper half of the Figure) has significantly lower
B-factors than the C-D dimer (lower half). This is a result of
the layered packing in the crystal lattice [Modis and Wierenga
1999]. Four molecules of acetyl-CoA are shown in ball-and-stick
representation, bound to each of the four subunits near the
interface between the A-B and C-D dimers. The side-chains of
residues located in high B-factor loops and pointing towards the
CoA moiety are also shown. These include residues Lys133,
Arg172, Lys208 and Arg232-Pro233-Ala234-Phe235-Asp236-Lys237,
which are all located in the loop domain. Figure 2, Figure 5 and
Figure 6 were prepared with MOLSCRIPT [Kraulis 1991] and
Raster3D [Merritt and Bacon 1997]. vert,
similar 5 Å2; red corresponds to 100 Å2. The A-B
dimer (upper half of the Figure) has significantly lower
B-factors than the C-D dimer (lower half). This is a result of
the layered packing in the crystal lattice [Modis and Wierenga
1999]. Four molecules of acetyl-CoA are shown in ball-and-stick
representation, bound to each of the four subunits near the
interface between the A-B and C-D dimers. The side-chains of
residues located in high B-factor loops and pointing towards the
CoA moiety are also shown. These include residues Lys133,
Arg172, Lys208 and Arg232-Pro233-Ala234-Phe235-Asp236-Lys237,
which are all located in the loop domain. Figure 2, Figure 5 and
Figure 6 were prepared with MOLSCRIPT [Kraulis 1991] and
Raster3D [Merritt and Bacon 1997].
|
 |
Figure 7.
Figure 7. Schematic drawing of the hydrogen bonding network
extending from the acitve site of biosynthetic thiolase, through
the enzyme to the molecular surface opposite the active site
pocket. Water molecules are shown as gray circles.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2000,
297,
1171-1182)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Pápai,
A.Hamza,
P.M.Pihko,
and
R.K.Wierenga
(2011).
Stereoelectronic requirements for optimal hydrogen-bond-catalyzed enolization.
|
| |
Chemistry,
17,
2859-2866.
|
 |
|
|
|
|
 |
D.E.Almonacid,
E.R.Yera,
J.B.Mitchell,
and
P.C.Babbitt
(2010).
Quantitative comparison of catalytic mechanisms and overall reactions in convergently evolved enzymes: implications for classification of enzyme function.
|
| |
PLoS Comput Biol,
6,
e1000700.
|
 |
|
|
|
|
 |
G.W.Han,
C.Bakolitsa,
M.D.Miller,
A.Kumar,
D.Carlton,
R.J.Najmanovich,
P.Abdubek,
T.Astakhova,
H.L.Axelrod,
C.Chen,
H.J.Chiu,
T.Clayton,
D.Das,
M.C.Deller,
L.Duan,
D.Ernst,
J.Feuerhelm,
J.C.Grant,
A.Grzechnik,
L.Jaroszewski,
K.K.Jin,
H.A.Johnson,
H.E.Klock,
M.W.Knuth,
P.Kozbial,
S.S.Krishna,
D.Marciano,
D.McMullan,
A.T.Morse,
E.Nigoghossian,
L.Okach,
R.Reyes,
C.L.Rife,
N.Sefcovic,
H.J.Tien,
C.B.Trame,
H.van den Bedem,
D.Weekes,
Q.Xu,
K.O.Hodgson,
J.Wooley,
M.A.Elsliger,
A.M.Deacon,
A.Godzik,
S.A.Lesley,
and
I.A.Wilson
(2010).
Structures of the first representatives of Pfam family PF06938 (DUF1285) reveal a new fold with repeated structural motifs and possible involvement in signal transduction.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
1218-1225.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Meriläinen,
W.Schmitz,
R.K.Wierenga,
and
P.Kursula
(2008).
The sulfur atoms of the substrate CoA and the catalytic cysteine are required for a productive mode of substrate binding in bacterial biosynthetic thiolase, a thioester-dependent enzyme.
|
| |
FEBS J,
275,
6136-6148.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.M.Haapalainen,
G.Meriläinen,
and
R.K.Wierenga
(2006).
The thiolase superfamily: condensing enzymes with diverse reaction specificities.
|
| |
Trends Biochem Sci,
31,
64-71.
|
 |
|
|
|
|
 |
Y.Meng,
and
J.Li
(2006).
Cloning, expression and characterization of a thiolase gene from Clostridium pasteurianum.
|
| |
Biotechnol Lett,
28,
1227-1232.
|
 |
|
|
|
|
 |
A.A.Pantazaki,
A.K.Ioannou,
and
D.A.Kyriakidis
(2005).
A thermostable beta-ketothiolase of polyhydroxyalkanoates (PHAs) in Thermus thermophilus: purification and biochemical properties.
|
| |
Mol Cell Biochem,
269,
27-36.
|
 |
|
|
|
|
 |
J.Peretó,
P.López-García,
and
D.Moreira
(2005).
Phylogenetic analysis of eukaryotic thiolases suggests multiple proteobacterial origins.
|
| |
J Mol Evol,
61,
65-74.
|
 |
|
|
|
|
 |
M.Ishikawa,
D.Tsuchiya,
T.Oyama,
Y.Tsunaka,
and
K.Morikawa
(2004).
Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex.
|
| |
EMBO J,
23,
2745-2754.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.J.Theisen,
I.Misra,
D.Saadat,
N.Campobasso,
H.M.Miziorko,
and
D.H.Harrison
(2004).
3-hydroxy-3-methylglutaryl-CoA synthase intermediate complex observed in "real-time".
|
| |
Proc Natl Acad Sci U S A,
101,
16442-16447.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.H.Dawe,
C.T.Porter,
J.M.Thornton,
and
A.B.Tabor
(2003).
A template search reveals mechanistic similarities and differences in beta-ketoacyl synthases (KAS) and related enzymes.
|
| |
Proteins,
52,
427-435.
|
 |
|
|
|
|
 |
T.Tsuge,
T.Hisano,
S.Taguchi,
and
Y.Doi
(2003).
Alteration of chain length substrate specificity of Aeromonas caviae R-enantiomer-specific enoyl-coenzyme A hydratase through site-directed mutagenesis.
|
| |
Appl Environ Microbiol,
69,
4830-4836.
|
 |
|
|
|
|
 |
C.M.Wilmot,
and
A.R.Pearson
(2002).
Cryocrystallography of metalloprotein reaction intermediates.
|
| |
Curr Opin Chem Biol,
6,
202-207.
|
 |
|
|
|
|
 |
M.Hedl,
A.Sutherlin,
E.I.Wilding,
M.Mazzulla,
D.McDevitt,
P.Lane,
J.W.Burgner,
K.R.Lehnbeuter,
C.V.Stauffacher,
M.N.Gwynn,
and
V.W.Rodwell
(2002).
Enterococcus faecalis acetoacetyl-coenzyme A thiolase/3-hydroxy-3-methylglutaryl-coenzyme A reductase, a dual-function protein of isopentenyl diphosphate biosynthesis.
|
| |
J Bacteriol,
184,
2116-2122.
|
 |
|
|
|
|
 |
T.Tsuge
(2002).
Metabolic improvements and use of inexpensive carbon sources in microbial production of polyhydroxyalkanoates.
|
| |
J Biosci Bioeng,
94,
579-584.
|
 |
|
|
|
|
 |
J.G.Olsen,
A.Kadziola,
P.von Wettstein-Knowles,
M.Siggaard-Andersen,
and
S.Larsen
(2001).
Structures of beta-ketoacyl-acyl carrier protein synthase I complexed with fatty acids elucidate its catalytic machinery.
|
| |
Structure,
9,
233-243.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Korotkova,
and
M.E.Lidstrom
(2001).
Connection between poly-beta-hydroxybutyrate biosynthesis and growth on C(1) and C(2) compounds in the methylotroph Methylobacterium extorquens AM1.
|
| |
J Bacteriol,
183,
1038-1046.
|
 |
|
|
|
|
 |
G.Taroncher-Oldenburg,
K.Nishina,
and
G.Stephanopoulos
(2000).
Identification and analysis of the polyhydroxyalkanoate-specific beta-ketothiolase and acetoacetyl coenzyme A reductase genes in the cyanobacterium Synechocystis sp. strain PCC6803.
|
| |
Appl Environ Microbiol,
66,
4440-4448.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

