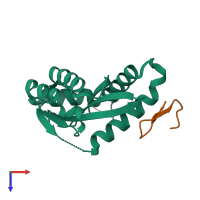
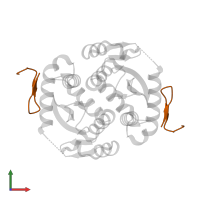
Assemblies
Assembly Name:
Protease and peptide
Multimeric state:
hetero tetramer
Accessible surface area:
13504.81 Å2
Buried surface area:
4111.68 Å2
Dissociation area:
647.07
Å2
Dissociation energy (ΔGdiss):
-8.03
kcal/mol
Dissociation entropy (TΔSdiss):
16.31
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-138021
Macromolecules
Chain: A
Length: 152 amino acids
Theoretical weight: 16.55 KDa
Source organism: Human immunodeficiency virus 1
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Integrase core domain
InterPro:
CATH: Ribonuclease H-like superfamily/Ribonuclease H
Length: 152 amino acids
Theoretical weight: 16.55 KDa
Source organism: Human immunodeficiency virus 1
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P04585 (Residues: 1204-1355; Coverage: 11%)
P04585 (Residues: 1204-1355; Coverage: 11%)
Pfam: Integrase core domain
InterPro:
CATH: Ribonuclease H-like superfamily/Ribonuclease H
Chain: B
Length: 15 amino acids
Theoretical weight: 2.01 KDa
Source organism: synthetic construct
Expression system: Not provided
Length: 15 amino acids
Theoretical weight: 2.01 KDa
Source organism: synthetic construct
Expression system: Not provided