Assemblies
Assembly Name:
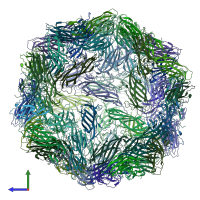
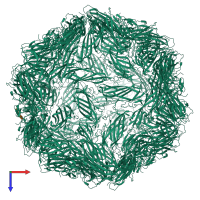
Cap protein
Multimeric state:
homo 60-mer
Accessible surface area:
407031.14 Å2
Buried surface area:
231437.48 Å2
Dissociation area:
115,311.19
Å2
Dissociation energy (ΔGdiss):
181.11
kcal/mol
Dissociation entropy (TΔSdiss):
836.01
kcal/mol
Symmetry number:
60
PDBe Complex ID:
PDB-CPX-182335
Macromolecules
Chains: A1, A2, A3, A4, A5, A6, A7, A8, A9, AA, AB, AC, AD, AE, AF, AG, AH, AI, AJ, AK, AL, AM, AN, AO, AP, AQ, AR, AS, AT, AU, AV, AW, AX, AY, AZ, Aa, Ab, Ac, Ad, Ae, Af, Ag, Ah, Ai, Aj, Ak, Al, Am, An, Ao, Ap, Aq, Ar, As, At, Au, Av, Aw, Ax, Ay
Length: 233 amino acids
Theoretical weight: 27.9 KDa
Source organism: Porcine circovirus 2
Expression system: Trichoplusia ni
UniProt:
Pfam: Circovirus capsid protein
InterPro:
Length: 233 amino acids
Theoretical weight: 27.9 KDa
Source organism: Porcine circovirus 2
Expression system: Trichoplusia ni
UniProt:
- Canonical:
 Q805N7 (Residues: 1-233; Coverage: 100%)
Q805N7 (Residues: 1-233; Coverage: 100%)
Pfam: Circovirus capsid protein
InterPro: