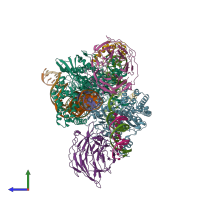
Assemblies
Multimeric state:
hetero undecamer
Accessible surface area:
115586.82 Å2
Buried surface area:
44141.76 Å2
Dissociation area:
1,515.71
Å2
Dissociation energy (ΔGdiss):
7.94
kcal/mol
Dissociation entropy (TΔSdiss):
13.71
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-119665
Macromolecules
Chains: A, C
Length: 776 amino acids
Theoretical weight: 88.67 KDa
Source organism: Mus musculus
Expression system: Homo sapiens
UniProt:
Pfam:
Length: 776 amino acids
Theoretical weight: 88.67 KDa
Source organism: Mus musculus
Expression system: Homo sapiens
UniProt:
- Canonical:
 P15919 (Residues: 265-1040; Coverage: 75%)
P15919 (Residues: 265-1040; Coverage: 75%)
Pfam:
- Zinc finger, C3HC4 type (RING finger)
- Recombination-activating protein 1 zinc-finger domain
- Recombination-activation protein 1 (RAG1), recombinase
Chain: N
Length: 163 amino acids
Theoretical weight: 18.9 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli K-12
UniProt:
Pfam:
InterPro:
Length: 163 amino acids
Theoretical weight: 18.9 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli K-12
UniProt:
- Canonical:
 P09429 (Residues: 1-163; Coverage: 76%)
P09429 (Residues: 1-163; Coverage: 76%)
Pfam:
InterPro:
Chains: B, D
Length: 520 amino acids
Theoretical weight: 58.16 KDa
Source organism: Mus musculus
Expression system: Homo sapiens
UniProt:
Pfam:
InterPro:
Length: 520 amino acids
Theoretical weight: 58.16 KDa
Source organism: Mus musculus
Expression system: Homo sapiens
UniProt:
- Canonical:
 P21784 (Residues: 1-520; Coverage: 99%)
P21784 (Residues: 1-520; Coverage: 99%)
Pfam:
InterPro:
Name:
DNA (5'-D(*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3')
Representative chains: I
Length: 16 nucleotides
Theoretical weight: 4.88 KDa
Representative chains: I
Length: 16 nucleotides
Theoretical weight: 4.88 KDa
Name:
DNA (5'-D(P*CP*TP*GP*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3')
Representative chains: J
Length: 20 nucleotides
Theoretical weight: 6.09 KDa
Representative chains: J
Length: 20 nucleotides
Theoretical weight: 6.09 KDa