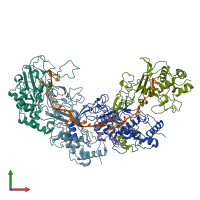
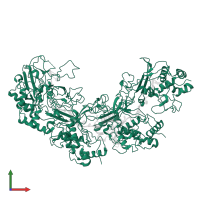
Assemblies
Assembly Name:
ACR30-35 and CRISPR-associated protein Csy3 and RNA
Multimeric state:
hetero hexamer
Accessible surface area:
64960.88 Å2
Buried surface area:
18050.65 Å2
Dissociation area:
931.53
Å2
Dissociation energy (ΔGdiss):
0.54
kcal/mol
Dissociation entropy (TΔSdiss):
11.65
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-125822
Macromolecules
Chains: C, D, E, F
Length: 342 amino acids
Theoretical weight: 37.58 KDa
Source organism: Pseudomonas aeruginosa UCBPP-PA14
Expression system: Escherichia coli 'BL21-Gold(DE3)pLysS AG'
UniProt:
Pfam: CRISPR-associated protein (Cas_Csy3)
InterPro: Type I-F CRISPR-associated protein Csy3
Length: 342 amino acids
Theoretical weight: 37.58 KDa
Source organism: Pseudomonas aeruginosa UCBPP-PA14
Expression system: Escherichia coli 'BL21-Gold(DE3)pLysS AG'
UniProt:
- Canonical:
 Q02MM1 (Residues: 1-342; Coverage: 100%)
Q02MM1 (Residues: 1-342; Coverage: 100%)
Pfam: CRISPR-associated protein (Cas_Csy3)
InterPro: Type I-F CRISPR-associated protein Csy3
Chain: M
Length: 78 amino acids
Theoretical weight: 8.82 KDa
Source organism: Pseudomonas phage JBD30
Expression system: Escherichia coli 'BL21-Gold(DE3)pLysS AG'
UniProt:
Pfam: Anti-CRISPR protein Acr30-35/AcrF1
Length: 78 amino acids
Theoretical weight: 8.82 KDa
Source organism: Pseudomonas phage JBD30
Expression system: Escherichia coli 'BL21-Gold(DE3)pLysS AG'
UniProt:
- Canonical:
 L7P7M1 (Residues: 1-78; Coverage: 100%)
L7P7M1 (Residues: 1-78; Coverage: 100%)
Pfam: Anti-CRISPR protein Acr30-35/AcrF1
Name:
crRNA with 20nt spacer sequence
Representative chains: K
Length: 48 nucleotides
Theoretical weight: 15.46 KDa
Representative chains: K
Length: 48 nucleotides
Theoretical weight: 15.46 KDa