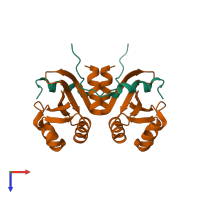
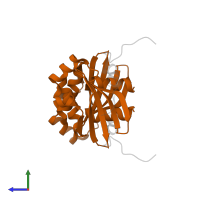
Assemblies
Multimeric state:
hetero tetramer
Accessible surface area:
12570.48 Å2
Buried surface area:
7097.23 Å2
Dissociation area:
1,676.02
Å2
Dissociation energy (ΔGdiss):
23.63
kcal/mol
Dissociation entropy (TΔSdiss):
12.02
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-150540
Macromolecules
Chain: B
Length: 26 amino acids
Theoretical weight: 2.89 KDa
Source organism: Saccharomyces cerevisiae S288C
Expression system: Escherichia coli
UniProt:
Length: 26 amino acids
Theoretical weight: 2.89 KDa
Source organism: Saccharomyces cerevisiae S288C
Expression system: Escherichia coli
UniProt:
- Canonical:
 P40537 (Residues: 821-845; Coverage: 2%)
P40537 (Residues: 821-845; Coverage: 2%)
Chain: A
Length: 122 amino acids
Theoretical weight: 14.14 KDa
Source organism: Saccharomyces cerevisiae S288C
Expression system: Escherichia coli
UniProt:
Pfam: Chromosome segregation protein Csm1/Pcs1
InterPro:
Length: 122 amino acids
Theoretical weight: 14.14 KDa
Source organism: Saccharomyces cerevisiae S288C
Expression system: Escherichia coli
UniProt:
- Canonical:
 P25651 (Residues: 69-190; Coverage: 64%)
P25651 (Residues: 69-190; Coverage: 64%)
Pfam: Chromosome segregation protein Csm1/Pcs1
InterPro:
- Monopolin complex subunit Csm1/Pcs1
- Monopolin complex subunit Csm1/Pcs1, C-terminal
- Csm1/Pcs1, C-terminal domain superfamily