Assemblies
Assembly Name:
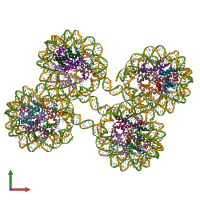
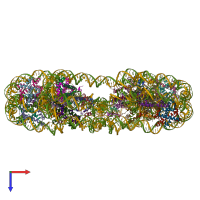
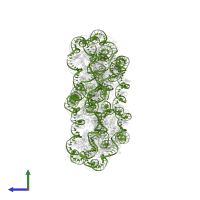
Nucleosome, Histone and DNA
Multimeric state:
hetero 34-mer
Accessible surface area:
298078.68 Å2
Buried surface area:
232793.12 Å2
Dissociation area:
4,322.73
Å2
Dissociation energy (ΔGdiss):
69.07
kcal/mol
Dissociation entropy (TΔSdiss):
12.99
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-119120
Macromolecules
Chains: D, H, L, P, T, X, b, f
Length: 126 amino acids
Theoretical weight: 13.98 KDa
Source organism: Xenopus laevis
Expression system: Escherichia coli
UniProt:
InterPro:
Length: 126 amino acids
Theoretical weight: 13.98 KDa
Source organism: Xenopus laevis
Expression system: Escherichia coli
UniProt:
- Canonical:
 P02281 (Residues: 1-126; Coverage: 100%)
P02281 (Residues: 1-126; Coverage: 100%)
InterPro:
Chains: A, E, I, M, Q, U, Y, c
Length: 135 amino acids
Theoretical weight: 15.3 KDa
Source organism: Xenopus laevis
Expression system: Escherichia coli
UniProt:
InterPro:
Length: 135 amino acids
Theoretical weight: 15.3 KDa
Source organism: Xenopus laevis
Expression system: Escherichia coli
UniProt:
- Canonical:
 P84233 (Residues: 2-136; Coverage: 99%)
P84233 (Residues: 2-136; Coverage: 99%)
InterPro:
Chains: B, F, J, N, R, V, Z, d
Length: 102 amino acids
Theoretical weight: 11.26 KDa
Source organism: Xenopus laevis
Expression system: Escherichia coli
UniProt:
InterPro:
Length: 102 amino acids
Theoretical weight: 11.26 KDa
Source organism: Xenopus laevis
Expression system: Escherichia coli
UniProt:
- Canonical:
 P62799 (Residues: 2-103; Coverage: 99%)
P62799 (Residues: 2-103; Coverage: 99%)
InterPro:
Chains: C, G, K, O, S, W, a, e
Length: 130 amino acids
Theoretical weight: 14.11 KDa
Source organism: Xenopus laevis
Expression system: Escherichia coli
UniProt:
InterPro:
Length: 130 amino acids
Theoretical weight: 14.11 KDa
Source organism: Xenopus laevis
Expression system: Escherichia coli
UniProt:
- Canonical:
 P06897 (Residues: 1-130; Coverage: 100%)
P06897 (Residues: 1-130; Coverage: 100%)
InterPro: