Assemblies
Assembly Name:
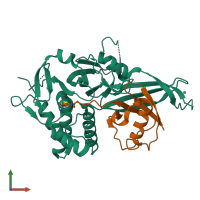
Ubiquitin carboxyl-terminal hydrolase 30 and Ubiquitin
Multimeric state:
hetero dimer
Accessible surface area:
16294.18 Å2
Buried surface area:
3162.37 Å2
Dissociation area:
1,581.19
Å2
Dissociation energy (ΔGdiss):
7.51
kcal/mol
Dissociation entropy (TΔSdiss):
11.36
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-143281
Assembly Name:
Ubiquitin carboxyl-terminal hydrolase 30 and Ubiquitin
Multimeric state:
hetero dimer
Accessible surface area:
16178.22 Å2
Buried surface area:
3155.02 Å2
Dissociation area:
1,577.51
Å2
Dissociation energy (ΔGdiss):
7.64
kcal/mol
Dissociation entropy (TΔSdiss):
11.36
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-143281
Macromolecules
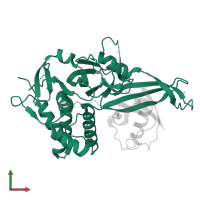
Chains: A, C
Length: 370 amino acids
Theoretical weight: 42.58 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Ubiquitin carboxyl-terminal hydrolase
InterPro:
Length: 370 amino acids
Theoretical weight: 42.58 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q70CQ3 (Residues: 64-357, 432-502; Coverage: 71%)
Q70CQ3 (Residues: 64-357, 432-502; Coverage: 71%)
Pfam: Ubiquitin carboxyl-terminal hydrolase
InterPro:
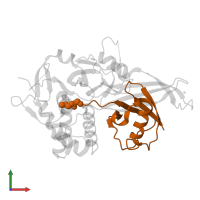
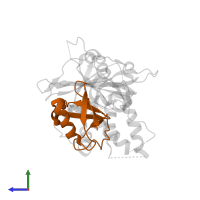
Chains: B, D
Length: 76 amino acids
Theoretical weight: 8.56 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Ubiquitin family
InterPro:
Length: 76 amino acids
Theoretical weight: 8.56 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P0CG47 (Residues: 153-228; Coverage: 33%)
P0CG47 (Residues: 153-228; Coverage: 33%)
Pfam: Ubiquitin family
InterPro: