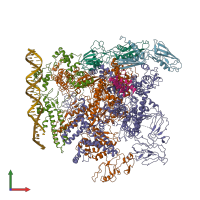
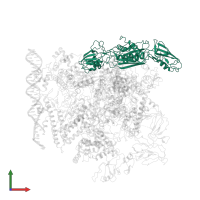
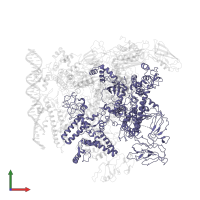
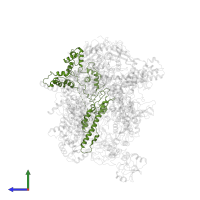
Assemblies
Multimeric state:
hetero octamer
Accessible surface area:
180138.04 Å2
Buried surface area:
31477.47 Å2
Dissociation area:
1,186.32
Å2
Dissociation energy (ΔGdiss):
5.6
kcal/mol
Dissociation entropy (TΔSdiss):
15.79
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-119071
Macromolecules
Chains: A, B
Length: 329 amino acids
Theoretical weight: 36.56 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam:
Length: 329 amino acids
Theoretical weight: 36.56 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P0A7Z4 (Residues: 1-329; Coverage: 100%)
P0A7Z4 (Residues: 1-329; Coverage: 100%)
Pfam:
- RNA polymerase Rpb3/Rpb11 dimerisation domain
- RNA polymerase Rpb3/RpoA insert domain
- Bacterial RNA polymerase, alpha chain C terminal domain
- DNA-directed RNA polymerase, alpha subunit
- RNA polymerase, RBP11-like subunit
- DNA-directed RNA polymerase, RpoA/D/Rpb3-type
- DNA-directed RNA polymerase, insert domain superfamily
- DNA-directed RNA polymerase, insert domain
- RNA polymerase, alpha subunit, C-terminal
Chain: C
Length: 1342 amino acids
Theoretical weight: 150.82 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam:
Length: 1342 amino acids
Theoretical weight: 150.82 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P0A8V2 (Residues: 1-1342; Coverage: 100%)
P0A8V2 (Residues: 1-1342; Coverage: 100%)
Pfam:
- RNA polymerase beta subunit
- RNA polymerase Rpb2, domain 2
- RNA polymerase Rpb2, domain 3
- RNA polymerase beta subunit external 1 domain
- RNA polymerase Rpb2, domain 6
- RNA polymerase Rpb2, domain 7
- DNA-directed RNA polymerase beta subunit, bacterial-type
- RNA polymerase, beta subunit, protrusion
- DNA-directed RNA polymerase, subunit 2
- RNA polymerase Rpb2, domain 2
- RNA polymerase Rpb2, domain 2 superfamily
- RNA polymerase Rpb2, domain 3
- DNA-directed RNA polymerase, beta subunit, external 1 domain superfamily
- DNA-directed RNA polymerase, beta subunit, external 1 domain
- DNA-directed RNA polymerase, subunit 2, hybrid-binding domain
- RNA polymerase Rpb2, OB-fold
- DNA-directed RNA polymerase, subunit 2, hybrid-binding domain superfamily
- RNA polymerase, beta subunit, conserved site
- RNA polymerase Rpb2, domain 7
Chain: D
Length: 1407 amino acids
Theoretical weight: 155.37 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam:
Length: 1407 amino acids
Theoretical weight: 155.37 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P0A8T7 (Residues: 1-1407; Coverage: 100%)
P0A8T7 (Residues: 1-1407; Coverage: 100%)
Pfam:
- RNA polymerase Rpb1, domain 1
- RNA polymerase Rpb1, domain 2
- RNA polymerase Rpb1, domain 3
- RNA polymerase Rpb1, domain 4
- RNA polymerase Rpb1, domain 5
- RNA polymerase Rpb1, clamp domain superfamily
- DNA-directed RNA polymerase, subunit beta-prime, bacterial type
- RNA polymerase Rpb1, domain 1
- DNA-directed RNA polymerase, subunit beta-prime
- RNA polymerase, N-terminal
- RNA polymerase, alpha subunit
- RNA polymerase Rpb1, domain 3
- RNA polymerase Rpb1, domain 3 superfamily
- RNA polymerase Rpb1, funnel domain superfamily
- RNA polymerase Rpb1, domain 4
- RNA polymerase Rpb1, domain 5
Chain: E
Length: 91 amino acids
Theoretical weight: 10.25 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: RNA polymerase Rpb6
InterPro:
Length: 91 amino acids
Theoretical weight: 10.25 KDa
Source organism: Escherichia coli K-12
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P0A800 (Residues: 1-91; Coverage: 100%)
P0A800 (Residues: 1-91; Coverage: 100%)
Pfam: RNA polymerase Rpb6
InterPro:
- DNA-directed RNA polymerase, omega subunit
- RPB6/omega subunit-like superfamily
- RNA polymerase, subunit omega/Rpo6/RPB6
Chain: M
Length: 573 amino acids
Theoretical weight: 62.16 KDa
Source organism: Klebsiella pneumoniae
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam:
Length: 573 amino acids
Theoretical weight: 62.16 KDa
Source organism: Klebsiella pneumoniae
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P06223 (Residues: 1-117, 118-305, 306-414, 415-477; Coverage: 100%)
P06223 (Residues: 1-117, 118-305, 306-414, 415-477; Coverage: 100%)
Pfam:
- Sigma-54 factor, Activator interacting domain (AID)
- Sigma-54 factor, core binding domain
- Sigma-54, DNA binding domain
Name:
Non-Template promoter DNA
Representative chains: F
Length: 63 nucleotides
Theoretical weight: 19.4 KDa
Representative chains: F
Length: 63 nucleotides
Theoretical weight: 19.4 KDa
Name:
Template DNA promoter
Representative chains: G
Length: 63 nucleotides
Theoretical weight: 19.44 KDa
Representative chains: G
Length: 63 nucleotides
Theoretical weight: 19.44 KDa