Assemblies
Assembly Name:
Eukaryotic translation initiation factor 4E
Multimeric state:
monomeric
Accessible surface area:
9491.7 Å2
Buried surface area:
277.09 Å2
Dissociation area:
138.55
Å2
Dissociation energy (ΔGdiss):
1.64
kcal/mol
Dissociation entropy (TΔSdiss):
0.86
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-158938
Assembly Name:
Eukaryotic translation initiation factor 4E
Multimeric state:
monomeric
Accessible surface area:
9464.09 Å2
Buried surface area:
264.03 Å2
Dissociation area:
132.01
Å2
Dissociation energy (ΔGdiss):
1.48
kcal/mol
Dissociation entropy (TΔSdiss):
0.86
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-158938
Assembly Name:
Eukaryotic translation initiation factor 4E
Multimeric state:
monomeric
Accessible surface area:
9280.47 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-158938
Assembly Name:
Eukaryotic translation initiation factor 4E
Multimeric state:
monomeric
Accessible surface area:
9389.45 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-158938
Macromolecules
Chains: A, B, C, D
Length: 190 amino acids
Theoretical weight: 22.15 KDa
Source organism: Mus musculus
Expression system: Escherichia coli
UniProt:
Pfam: Eukaryotic initiation factor 4E
InterPro:
Length: 190 amino acids
Theoretical weight: 22.15 KDa
Source organism: Mus musculus
Expression system: Escherichia coli
UniProt:
- Canonical:
 P63073 (Residues: 28-217; Coverage: 88%)
P63073 (Residues: 28-217; Coverage: 88%)
Pfam: Eukaryotic initiation factor 4E
InterPro:
- Translation Initiation factor eIF- 4e-like
- Translation Initiation factor eIF- 4e
- Eukaryotic translation initiation factor 4E (eIF-4E), conserved site



![The deposited structure of PDB entry <span class='highlight'>5m7x</span> coloured by chain, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>4 copies of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>4 copies of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>GLYCEROL</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 5m7x coloured by chain, front view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_deposited_chain_front_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>5m7x</span> coloured by chain, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>4 copies of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>4 copies of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>GLYCEROL</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 5m7x coloured by chain, side view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_deposited_chain_side_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>5m7x</span> coloured by chain, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>4 copies of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>4 copies of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>GLYCEROL</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 5m7x coloured by chain, top view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_deposited_chain_top_image-200x200.png)
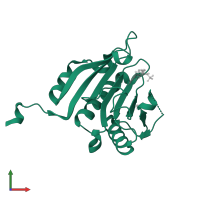
![Monomeric assembly 1 of PDB entry <span class='highlight'>5m7x</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>GLYCEROL</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 1 of PDB entry 5m7x coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_assembly_1_chemically_distinct_molecules_front_image-200x200.png)
![Monomeric assembly 1 of PDB entry <span class='highlight'>5m7x</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>GLYCEROL</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 1 of PDB entry 5m7x coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_assembly_1_chemically_distinct_molecules_side_image-200x200.png)
![Monomeric assembly 1 of PDB entry <span class='highlight'>5m7x</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>GLYCEROL</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 1 of PDB entry 5m7x coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_assembly_1_chemically_distinct_molecules_top_image-200x200.png)
![Monomeric assembly 2 of PDB entry <span class='highlight'>5m7x</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>GLYCEROL</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 2 of PDB entry 5m7x coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_assembly_2_chemically_distinct_molecules_front_image-200x200.png)
![Monomeric assembly 2 of PDB entry <span class='highlight'>5m7x</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>GLYCEROL</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 2 of PDB entry 5m7x coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_assembly_2_chemically_distinct_molecules_side_image-200x200.png)
![Monomeric assembly 2 of PDB entry <span class='highlight'>5m7x</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>GLYCEROL</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 2 of PDB entry 5m7x coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_assembly_2_chemically_distinct_molecules_top_image-200x200.png)
![Monomeric assembly 3 of PDB entry <span class='highlight'>5m7x</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 3 of PDB entry 5m7x coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_assembly_3_chemically_distinct_molecules_front_image-200x200.png)
![Monomeric assembly 3 of PDB entry <span class='highlight'>5m7x</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 3 of PDB entry 5m7x coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_assembly_3_chemically_distinct_molecules_side_image-200x200.png)
![Monomeric assembly 3 of PDB entry <span class='highlight'>5m7x</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 3 of PDB entry 5m7x coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_assembly_3_chemically_distinct_molecules_top_image-200x200.png)
![Monomeric assembly 4 of PDB entry <span class='highlight'>5m7x</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 4 of PDB entry 5m7x coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_assembly_4_chemically_distinct_molecules_front_image-200x200.png)
![Monomeric assembly 4 of PDB entry <span class='highlight'>5m7x</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 4 of PDB entry 5m7x coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_assembly_4_chemically_distinct_molecules_side_image-200x200.png)
![Monomeric assembly 4 of PDB entry <span class='highlight'>5m7x</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Eukaryotic translation initiation factor 4E</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>[(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 4 of PDB entry 5m7x coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/5m7x_assembly_4_chemically_distinct_molecules_top_image-200x200.png)