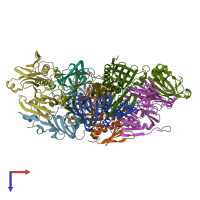
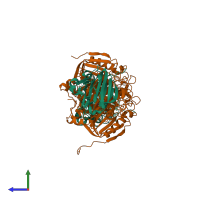
Assemblies
Assembly Name:
Hydroquinone dioxygenase small subunit and Hydroquinone 1,2-dioxygenase large subunit N-terminal domain-containing protein
Multimeric state:
hetero tetramer
Accessible surface area:
34538.18 Å2
Buried surface area:
16336.63 Å2
Dissociation area:
1,828.95
Å2
Dissociation energy (ΔGdiss):
14.72
kcal/mol
Dissociation entropy (TΔSdiss):
14.78
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-123731
Assembly Name:
Hydroquinone dioxygenase small subunit and Hydroquinone 1,2-dioxygenase large subunit N-terminal domain-containing protein
Multimeric state:
hetero tetramer
Accessible surface area:
34538.18 Å2
Buried surface area:
16336.63 Å2
Dissociation area:
1,828.95
Å2
Dissociation energy (ΔGdiss):
14.72
kcal/mol
Dissociation entropy (TΔSdiss):
14.78
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-123731
Assembly Name:
Hydroquinone dioxygenase small subunit and Hydroquinone 1,2-dioxygenase large subunit N-terminal domain-containing protein
Multimeric state:
hetero tetramer
Accessible surface area:
35225.19 Å2
Buried surface area:
15740.31 Å2
Dissociation area:
1,825.81
Å2
Dissociation energy (ΔGdiss):
15.28
kcal/mol
Dissociation entropy (TΔSdiss):
14.79
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-123731
Macromolecules
Chains: A, C, E, G
Length: 170 amino acids
Theoretical weight: 18.57 KDa
Source organism: Sphingomonas sp. TTNP3
UniProt:
Length: 170 amino acids
Theoretical weight: 18.57 KDa
Source organism: Sphingomonas sp. TTNP3
UniProt:
- Canonical:
 F8TW82 (Residues: 1-170; Coverage: 100%)
F8TW82 (Residues: 1-170; Coverage: 100%)
Chains: B, D, F, H
Length: 341 amino acids
Theoretical weight: 38.24 KDa
Source organism: Sphingomonas sp. TTNP3
UniProt:
Pfam: Hydroquinone 1,2-dioxygenase large subunit N-terminal
InterPro:
Length: 341 amino acids
Theoretical weight: 38.24 KDa
Source organism: Sphingomonas sp. TTNP3
UniProt:
- Canonical:
 F8TW83 (Residues: 1-341; Coverage: 100%)
F8TW83 (Residues: 1-341; Coverage: 100%)
Pfam: Hydroquinone 1,2-dioxygenase large subunit N-terminal
InterPro: