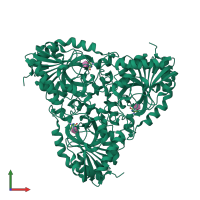
Assemblies
Assembly Name:
Purine nucleoside phosphorylase
Multimeric state:
homo trimer
Accessible surface area:
30341.6 Å2
Buried surface area:
9821.95 Å2
Dissociation area:
3,816.93
Å2
Dissociation energy (ΔGdiss):
49.96
kcal/mol
Dissociation entropy (TΔSdiss):
27.58
kcal/mol
Symmetry number:
3
PDBe Complex ID:
PDB-CPX-102740
Macromolecules
Chain: A
Length: 287 amino acids
Theoretical weight: 31.36 KDa
Source organism: Schistosoma mansoni
Expression system: Escherichia coli
UniProt:
InterPro:
Length: 287 amino acids
Theoretical weight: 31.36 KDa
Source organism: Schistosoma mansoni
Expression system: Escherichia coli
UniProt:
- Canonical:
 A0A0U3AGT1 (Residues: 1-287; Coverage: 100%)
A0A0U3AGT1 (Residues: 1-287; Coverage: 100%)
InterPro:
- Purine nucleoside phosphorylase
- Nucleoside phosphorylase superfamily
- Nucleoside phosphorylase domain
- Purine nucleoside phosphorylase I, inosine/guanosine-specific