Assemblies
Assembly Name:
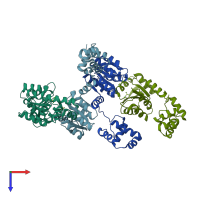
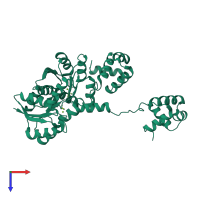
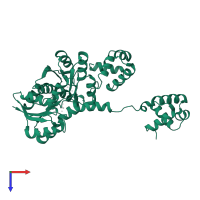
Response regulator protein VraR
Multimeric state:
homo dimer
Accessible surface area:
22087.04 Å2
Buried surface area:
2077.61 Å2
Dissociation area:
719.74
Å2
Dissociation energy (ΔGdiss):
2.21
kcal/mol
Dissociation entropy (TΔSdiss):
12.89
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-197411
Assembly Name:
Response regulator protein VraR
Multimeric state:
homo dimer
Accessible surface area:
22266.8 Å2
Buried surface area:
2134.19 Å2
Dissociation area:
750.37
Å2
Dissociation energy (ΔGdiss):
1.85
kcal/mol
Dissociation entropy (TΔSdiss):
12.91
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-197411
Macromolecules
Chains: A, B, C, F
Length: 210 amino acids
Theoretical weight: 23.68 KDa
Source organism: Enterococcus faecium SD3B-2
Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
Length: 210 amino acids
Theoretical weight: 23.68 KDa
Source organism: Enterococcus faecium SD3B-2
Expression system: Escherichia coli
UniProt:
- Canonical:
 S4FAU8 (Residues: 1-210; Coverage: 99%)
S4FAU8 (Residues: 1-210; Coverage: 99%)
Pfam:
InterPro: