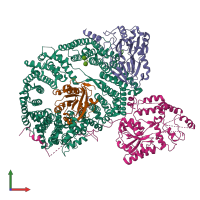
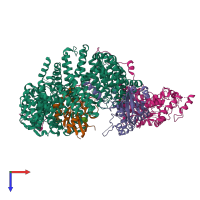
Assemblies
Multimeric state:
hetero tetramer
Accessible surface area:
80915.51 Å2
Buried surface area:
15233.59 Å2
Dissociation area:
2,228.03
Å2
Dissociation energy (ΔGdiss):
3.65
kcal/mol
Dissociation entropy (TΔSdiss):
13.71
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-127257
Macromolecules
Chain: A
Length: 1044 amino acids
Theoretical weight: 120.4 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam:
Length: 1044 amino acids
Theoretical weight: 120.4 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 O14980 (Residues: 5-1048; Coverage: 98%)
O14980 (Residues: 5-1048; Coverage: 98%)
Pfam:
- Importin-beta N-terminal domain
- Exportin 1-like protein
- Chromosome region maintenance or exportin repeat
- CRM1 / Exportin repeat 2
- CRM1 / Exportin repeat 3
- CRM1 C terminal
Chain: B
Length: 172 amino acids
Theoretical weight: 19.81 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Ras family
InterPro:
Length: 172 amino acids
Theoretical weight: 19.81 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P62826 (Residues: 8-179; Coverage: 80%)
P62826 (Residues: 8-179; Coverage: 80%)
Pfam: Ras family
InterPro:
- Small GTPase
- Ran GTPase
- P-loop containing nucleoside triphosphate hydrolase
- Small GTP-binding protein domain
Chain: C
Length: 289 amino acids
Theoretical weight: 33.28 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Snurportin1
InterPro:
Length: 289 amino acids
Theoretical weight: 33.28 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 O95149 (Residues: 1-287; Coverage: 80%)
O95149 (Residues: 1-287; Coverage: 80%)
Pfam: Snurportin1
InterPro:
- Importin-alpha, importin-beta-binding domain
- Snurportin-1
- Snurportin-1, N-terminal
- Snurportin-1, m3G cap-binding domain
Chain: D
Length: 479 amino acids
Theoretical weight: 50.58 KDa
Source organisms: Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
Length: 479 amino acids
Theoretical weight: 50.58 KDa
Source organisms: Expression system: Escherichia coli
UniProt:
- Canonical:
 P0AEX9 (Residues: 32-387; Coverage: 96%)
P0AEX9 (Residues: 32-387; Coverage: 96%) - Canonical:
 P35658 (Residues: 1916-2027; Coverage: 5%)
nullnull
P35658 (Residues: 1916-2027; Coverage: 5%)
nullnull
Pfam:
InterPro: