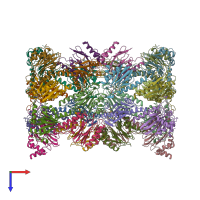
Assemblies
Assembly Name:
20S Proteasome complex
Multimeric state:
hetero 28-mer
Accessible surface area:
202461.85 Å2
Buried surface area:
86759.04 Å2
Dissociation area:
6,149.99
Å2
Dissociation energy (ΔGdiss):
47.83
kcal/mol
Dissociation entropy (TΔSdiss):
28.32
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-127173
Macromolecules
Chains: A, O
Length: 246 amino acids
Theoretical weight: 27.43 KDa
Source organism: Homo sapiens
UniProt:
Pfam:
InterPro:
Length: 246 amino acids
Theoretical weight: 27.43 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 P60900 (Residues: 1-246; Coverage: 100%)
P60900 (Residues: 1-246; Coverage: 100%)
Pfam:
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome subunit alpha6
- Proteasome alpha-subunit, N-terminal domain
- Proteasome alpha-type subunit
- Proteasome, subunit alpha/beta
Chains: B, P
Length: 234 amino acids
Theoretical weight: 25.93 KDa
Source organism: Homo sapiens
UniProt:
Pfam:
InterPro:
Length: 234 amino acids
Theoretical weight: 25.93 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 P25787 (Residues: 1-234; Coverage: 100%)
P25787 (Residues: 1-234; Coverage: 100%)
Pfam:
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome alpha-subunit, N-terminal domain
- Proteasome alpha-type subunit
- Proteasome, subunit alpha/beta
Chains: C, Q
Length: 261 amino acids
Theoretical weight: 29.53 KDa
Source organism: Homo sapiens
UniProt:
Pfam:
InterPro:
Length: 261 amino acids
Theoretical weight: 29.53 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 P25789 (Residues: 1-261; Coverage: 100%)
P25789 (Residues: 1-261; Coverage: 100%)
Pfam:
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome alpha-subunit, N-terminal domain
- Proteasome alpha-type subunit
- Proteasome, subunit alpha/beta
- Proteasome beta-type subunit, conserved site
Chains: D, R
Length: 248 amino acids
Theoretical weight: 27.93 KDa
Source organism: Homo sapiens
UniProt:
Pfam:
InterPro:
Length: 248 amino acids
Theoretical weight: 27.93 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 O14818 (Residues: 1-248; Coverage: 100%)
O14818 (Residues: 1-248; Coverage: 100%)
Pfam:
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome alpha-subunit, N-terminal domain
- Proteasome alpha-type subunit
- Proteasome, subunit alpha/beta
Chains: E, S
Length: 241 amino acids
Theoretical weight: 26.44 KDa
Source organism: Homo sapiens
UniProt:
Pfam:
InterPro:
Length: 241 amino acids
Theoretical weight: 26.44 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 P28066 (Residues: 1-241; Coverage: 100%)
P28066 (Residues: 1-241; Coverage: 100%)
Pfam:
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome alpha-subunit, N-terminal domain
- Proteasome alpha-type subunit
- Proteasome, subunit alpha/beta
- Proteasome subunit alpha5
Chains: F, T
Length: 263 amino acids
Theoretical weight: 29.6 KDa
Source organism: Homo sapiens
UniProt:
Pfam:
InterPro:
Length: 263 amino acids
Theoretical weight: 29.6 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 P25786 (Residues: 1-263; Coverage: 100%)
P25786 (Residues: 1-263; Coverage: 100%)
Pfam:
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome alpha-subunit, N-terminal domain
- Proteasome alpha-type subunit
- Proteasome, subunit alpha/beta
- Proteasome subunit alpha 1
Chains: G, U
Length: 255 amino acids
Theoretical weight: 28.47 KDa
Source organism: Homo sapiens
UniProt:
Pfam:
InterPro:
Length: 255 amino acids
Theoretical weight: 28.47 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 P25788 (Residues: 1-255; Coverage: 100%)
P25788 (Residues: 1-255; Coverage: 100%)
Pfam:
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome alpha-subunit, N-terminal domain
- Proteasome alpha-type subunit
- Proteasome, subunit alpha/beta
Chains: H, V
Length: 205 amino acids
Theoretical weight: 21.92 KDa
Source organism: Homo sapiens
UniProt:
Pfam: Proteasome subunit
InterPro:
Length: 205 amino acids
Theoretical weight: 21.92 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 P28072 (Residues: 35-239; Coverage: 86%)
P28072 (Residues: 35-239; Coverage: 86%)
Pfam: Proteasome subunit
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome, subunit alpha/beta
- Proteasome beta-type subunit, conserved site
- Proteasome B-type subunit
- Peptidase T1A, proteasome beta-subunit
Chains: I, W
Length: 234 amino acids
Theoretical weight: 25.32 KDa
Source organism: Homo sapiens
UniProt:
Pfam:
InterPro:
Length: 234 amino acids
Theoretical weight: 25.32 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 Q99436 (Residues: 44-277; Coverage: 85%)
Q99436 (Residues: 44-277; Coverage: 85%)
Pfam:
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome, subunit alpha/beta
- Proteasome beta-type subunit, conserved site
- Proteasome B-type subunit
- Peptidase T1A, proteasome beta-subunit
- Proteasome beta subunit, C-terminal
Chains: J, X
Length: 204 amino acids
Theoretical weight: 22.84 KDa
Source organism: Homo sapiens
UniProt:
Pfam: Proteasome subunit
InterPro:
Length: 204 amino acids
Theoretical weight: 22.84 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 P49720 (Residues: 2-205; Coverage: 100%)
P49720 (Residues: 2-205; Coverage: 100%)
Pfam: Proteasome subunit
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome, subunit alpha/beta
- Proteasome beta-type subunit, conserved site
- Proteasome B-type subunit
- Proteasome beta 3 subunit
Chains: K, Y
Length: 201 amino acids
Theoretical weight: 22.86 KDa
Source organism: Homo sapiens
UniProt:
Pfam: Proteasome subunit
InterPro:
Length: 201 amino acids
Theoretical weight: 22.86 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 P49721 (Residues: 1-201; Coverage: 100%)
P49721 (Residues: 1-201; Coverage: 100%)
Pfam: Proteasome subunit
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome, subunit alpha/beta
- Proteasome beta-type subunit, conserved site
- Proteasome B-type subunit
- Proteasome subunit beta 2
Chains: L, Z
Length: 204 amino acids
Theoretical weight: 22.48 KDa
Source organism: Homo sapiens
UniProt:
Pfam: Proteasome subunit
InterPro:
Length: 204 amino acids
Theoretical weight: 22.48 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 P28074 (Residues: 60-263; Coverage: 78%)
P28074 (Residues: 60-263; Coverage: 78%)
Pfam: Proteasome subunit
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome, subunit alpha/beta
- Proteasome beta-type subunit, conserved site
- Proteasome B-type subunit
- Peptidase T1A, proteasome beta-subunit
Chains: M, a
Length: 213 amino acids
Theoretical weight: 23.58 KDa
Source organism: Homo sapiens
UniProt:
Pfam: Proteasome subunit
InterPro:
Length: 213 amino acids
Theoretical weight: 23.58 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 P20618 (Residues: 29-241; Coverage: 88%)
P20618 (Residues: 29-241; Coverage: 88%)
Pfam: Proteasome subunit
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome, subunit alpha/beta
- Proteasome beta-type subunit, conserved site
- Proteasome B-type subunit
Chains: N, b
Length: 219 amino acids
Theoretical weight: 24.41 KDa
Source organism: Homo sapiens
UniProt:
Pfam: Proteasome subunit
InterPro:
Length: 219 amino acids
Theoretical weight: 24.41 KDa
Source organism: Homo sapiens
UniProt:
- Canonical:
 P28070 (Residues: 46-264; Coverage: 83%)
P28070 (Residues: 46-264; Coverage: 83%)
Pfam: Proteasome subunit
InterPro:
- Nucleophile aminohydrolases, N-terminal
- Proteasome, subunit alpha/beta
- Proteasome beta-type subunit, conserved site
- Proteasome B-type subunit
- Proteasome subunit beta 4