Assemblies
Assembly Name:
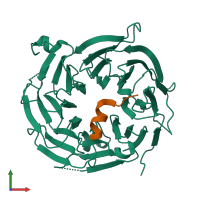
60S ribosomal protein L10-like protein and sqt1
Multimeric state:
hetero dimer
Accessible surface area:
15707.5 Å2
Buried surface area:
1369.74 Å2
Dissociation area:
684.87
Å2
Dissociation energy (ΔGdiss):
3.2
kcal/mol
Dissociation entropy (TΔSdiss):
7.92
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-123947
Assembly Name:
60S ribosomal protein L10-like protein and sqt1
Multimeric state:
hetero dimer
Accessible surface area:
15598.42 Å2
Buried surface area:
1401.33 Å2
Dissociation area:
700.67
Å2
Dissociation energy (ΔGdiss):
2.85
kcal/mol
Dissociation entropy (TΔSdiss):
7.91
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-123947
Macromolecules
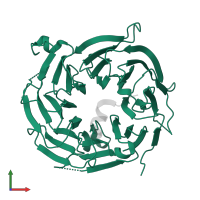
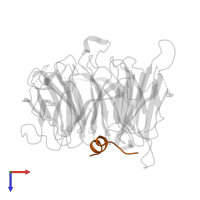
Chains: A, B
Length: 480 amino acids
Theoretical weight: 50.3 KDa
Source organism: Thermochaetoides thermophila
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: WD domain, G-beta repeat
InterPro:
Length: 480 amino acids
Theoretical weight: 50.3 KDa
Source organism: Thermochaetoides thermophila
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 G0S0R0 (Residues: 54-533; Coverage: 90%)
G0S0R0 (Residues: 54-533; Coverage: 90%)
Pfam: WD domain, G-beta repeat
InterPro:
- WD40 repeat
- WD40/YVTN repeat-like-containing domain superfamily
- Soluble quinoprotein glucose/sorbosone dehydrogenase
- WD40-repeat-containing domain superfamily
Chains: C, Y
Length: 28 amino acids
Theoretical weight: 3.47 KDa
Source organism: Thermochaetoides thermophila
Expression system: Escherichia coli BL21(DE3)
UniProt:
InterPro: Large ribosomal subunit protein uL16 superfamily
Length: 28 amino acids
Theoretical weight: 3.47 KDa
Source organism: Thermochaetoides thermophila
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 G0SEI1 (Residues: 1-20; Coverage: 9%)
G0SEI1 (Residues: 1-20; Coverage: 9%)
InterPro: Large ribosomal subunit protein uL16 superfamily