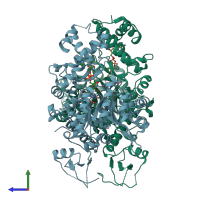
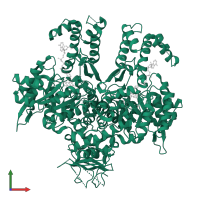
Assemblies
Assembly Name:
Omega-3 polyunsaturated fatty acid synthase subunit PfaD
Multimeric state:
homo dimer
Accessible surface area:
38738.64 Å2
Buried surface area:
7204.06 Å2
Dissociation area:
1,535.19
Å2
Dissociation energy (ΔGdiss):
13.36
kcal/mol
Dissociation entropy (TΔSdiss):
14.95
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-184390
Macromolecules
Chains: A, B
Length: 547 amino acids
Theoretical weight: 59.67 KDa
Source organism: Shewanella oneidensis MR-1
Expression system: Escherichia coli
UniProt:
Pfam:
Length: 547 amino acids
Theoretical weight: 59.67 KDa
Source organism: Shewanella oneidensis MR-1
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q8EGK4 (Residues: 1-547; Coverage: 100%)
Q8EGK4 (Residues: 1-547; Coverage: 100%)
Pfam:
- Fatty acid synthase subunit PfaD N-terminal domain
- Nitronate monooxygenase
- [Acyl-carrier-protein] S-malonyltransferase, inserted helical domain