Assemblies
Assembly Name:
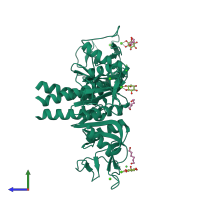
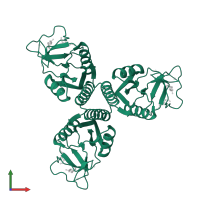
Pulmonary surfactant-associated protein A
Multimeric state:
homo trimer
Accessible surface area:
19343.8 Å2
Buried surface area:
6892.35 Å2
Dissociation area:
2,150.56
Å2
Dissociation energy (ΔGdiss):
25.93
kcal/mol
Dissociation entropy (TΔSdiss):
24.49
kcal/mol
Symmetry number:
3
PDBe Complex ID:
PDB-CPX-140125
Macromolecules
Chain: A
Length: 148 amino acids
Theoretical weight: 16.63 KDa
Source organism: Rattus norvegicus
Expression system: Trichoplusia ni
UniProt:
Pfam: Lectin C-type domain
InterPro:
Length: 148 amino acids
Theoretical weight: 16.63 KDa
Source organism: Rattus norvegicus
Expression system: Trichoplusia ni
UniProt:
- Canonical:
 P08427 (Residues: 101-248; Coverage: 65%)
P08427 (Residues: 101-248; Coverage: 65%)
Pfam: Lectin C-type domain
InterPro:
- C-type lectin-like/link domain superfamily
- C-type lectin-like
- C-type lectin fold
- Collectin, C-type lectin-like domain
- C-type lectin, conserved site