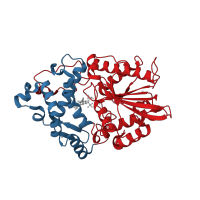
The Complex Structure of Mutant Phytase with IHS
Source organism:
Escherichia coli
Biochemical function:
inositol phosphate phosphatase activity
Biological process: cellular response to anoxia
Cellular component: periplasmic space
Biological process: cellular response to anoxia
Cellular component: periplasmic space