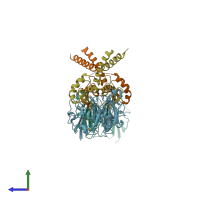
Assemblies
Multimeric state:
hetero tetramer
Accessible surface area:
38999.35 Å2
Buried surface area:
5618.9 Å2
Dissociation area:
1,203.54
Å2
Dissociation energy (ΔGdiss):
-19.28
kcal/mol
Dissociation entropy (TΔSdiss):
26.3
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-142728
Macromolecules
Chains: A, B
Length: 259 amino acids
Theoretical weight: 28.3 KDa
Source organism: Escherichia coli str. K-12 substr. MG1655
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: DNA gyrase C-terminal domain, beta-propeller
InterPro:
Length: 259 amino acids
Theoretical weight: 28.3 KDa
Source organism: Escherichia coli str. K-12 substr. MG1655
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P0AFI2 (Residues: 497-752; Coverage: 34%)
P0AFI2 (Residues: 497-752; Coverage: 34%)
Pfam: DNA gyrase C-terminal domain, beta-propeller
InterPro:
Chains: C, D
Length: 163 amino acids
Theoretical weight: 18.35 KDa
Source organism: Escherichia coli str. K-12 substr. MG1655
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: MukB hinge domain
InterPro:
CATH:
Length: 163 amino acids
Theoretical weight: 18.35 KDa
Source organism: Escherichia coli str. K-12 substr. MG1655
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P22523 (Residues: 645-804; Coverage: 11%)
P22523 (Residues: 645-804; Coverage: 11%)
Pfam: MukB hinge domain
InterPro:
CATH: