Assemblies
Assembly Name:
Heme NO-binding domain-containing protein
Multimeric state:
monomeric
Accessible surface area:
9098.0 Å2
Buried surface area:
255.3 Å2
Dissociation area:
127.65
Å2
Dissociation energy (ΔGdiss):
-2.49
kcal/mol
Dissociation entropy (TΔSdiss):
1.21
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-187121
Assembly Name:
Heme NO-binding domain-containing protein
Multimeric state:
monomeric
Accessible surface area:
9002.79 Å2
Buried surface area:
274.51 Å2
Dissociation area:
137.25
Å2
Dissociation energy (ΔGdiss):
-2.51
kcal/mol
Dissociation entropy (TΔSdiss):
1.2
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-187121
Assembly Name:
Heme NO-binding domain-containing protein
Multimeric state:
homo trimer
Accessible surface area:
23628.32 Å2
Buried surface area:
4409.57 Å2
Dissociation area:
1,820.6
Å2
Dissociation energy (ΔGdiss):
2.38
kcal/mol
Dissociation entropy (TΔSdiss):
25.43
kcal/mol
Symmetry number:
3
PDBe Complex ID:
PDB-CPX-187123
Assembly Name:
Heme NO-binding domain-containing protein
Multimeric state:
homo trimer
Accessible surface area:
23259.03 Å2
Buried surface area:
4565.96 Å2
Dissociation area:
1,872.8
Å2
Dissociation energy (ΔGdiss):
5.89
kcal/mol
Dissociation entropy (TΔSdiss):
25.42
kcal/mol
Symmetry number:
3
PDBe Complex ID:
PDB-CPX-187123
Assembly Name:
Heme NO-binding domain-containing protein
Multimeric state:
homo hexamer
Accessible surface area:
43971.27 Å2
Buried surface area:
11904.42 Å2
Dissociation area:
5,451.74
Å2
Dissociation energy (ΔGdiss):
-0.21
kcal/mol
Dissociation entropy (TΔSdiss):
65.6
kcal/mol
Symmetry number:
6
PDBe Complex ID:
PDB-CPX-187124
Macromolecules
Chains: A, B
Length: 182 amino acids
Theoretical weight: 20.39 KDa
Source organism: Nostoc sp. PCC 7120 = FACHB-418
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Haem-NO-binding
InterPro:
Length: 182 amino acids
Theoretical weight: 20.39 KDa
Source organism: Nostoc sp. PCC 7120 = FACHB-418
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q8YUQ7 (Residues: 1-182; Coverage: 96%)
Q8YUQ7 (Residues: 1-182; Coverage: 96%)
Pfam: Haem-NO-binding
InterPro:
- NO signalling/Golgi transport ligand-binding domain superfamily
- H-NOX domain superfamily
- Heme NO-binding



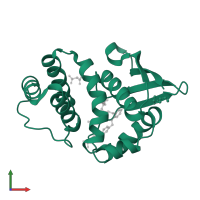
![The deposited structure of PDB entry <span class='highlight'>4iah</span> coloured by chain, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 4iah coloured by chain, front view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_deposited_chain_front_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>4iah</span> coloured by chain, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 4iah coloured by chain, side view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_deposited_chain_side_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>4iah</span> coloured by chain, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>2 copies of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>2 copies of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 4iah coloured by chain, top view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_deposited_chain_top_image-200x200.png)
![Monomeric assembly 1 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 1 of PDB entry 4iah coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_1_chemically_distinct_molecules_front_image-200x200.png)
![Monomeric assembly 1 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 1 of PDB entry 4iah coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_1_chemically_distinct_molecules_side_image-200x200.png)
![Monomeric assembly 1 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 1 of PDB entry 4iah coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_1_chemically_distinct_molecules_top_image-200x200.png)
![Monomeric assembly 2 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 2 of PDB entry 4iah coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_2_chemically_distinct_molecules_front_image-200x200.png)
![Monomeric assembly 2 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 2 of PDB entry 4iah coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_2_chemically_distinct_molecules_side_image-200x200.png)
![Monomeric assembly 2 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 2 of PDB entry 4iah coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_2_chemically_distinct_molecules_top_image-200x200.png)
![Homo trimeric assembly 3 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>3 copies of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>water</span>.</li></ul>} Homo trimeric assembly 3 of PDB entry 4iah coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_3_chemically_distinct_molecules_front_image-200x200.png)
![Homo trimeric assembly 3 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>3 copies of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>water</span>.</li></ul>} Homo trimeric assembly 3 of PDB entry 4iah coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_3_chemically_distinct_molecules_side_image-200x200.png)
![Homo trimeric assembly 3 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>3 copies of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>water</span>.</li></ul>} Homo trimeric assembly 3 of PDB entry 4iah coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_3_chemically_distinct_molecules_top_image-200x200.png)
![Homo trimeric assembly 4 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>3 copies of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>water</span>.</li></ul>} Homo trimeric assembly 4 of PDB entry 4iah coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_4_chemically_distinct_molecules_front_image-200x200.png)
![Homo trimeric assembly 4 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>3 copies of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>water</span>.</li></ul>} Homo trimeric assembly 4 of PDB entry 4iah coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_4_chemically_distinct_molecules_side_image-200x200.png)
![Homo trimeric assembly 4 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>3 copies of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>water</span>.</li></ul>} Homo trimeric assembly 4 of PDB entry 4iah coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_4_chemically_distinct_molecules_top_image-200x200.png)
![Homo hexameric assembly 5 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>6 copies of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>water</span>.</li></ul>} Homo hexameric assembly 5 of PDB entry 4iah coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_5_chemically_distinct_molecules_front_image-200x200.png)
![Homo hexameric assembly 5 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>6 copies of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>water</span>.</li></ul>} Homo hexameric assembly 5 of PDB entry 4iah coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_5_chemically_distinct_molecules_side_image-200x200.png)
![Homo hexameric assembly 5 of PDB entry <span class='highlight'>4iah</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>6 copies of <span class='highlight'>Heme NO-binding domain-containing protein</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid</span>;</li> <li class='image_legend_li'>6 copies of <span class='highlight'>MALONATE ION</span>;</li> <li class='image_legend_li'>3 copies of <span class='highlight'>water</span>.</li></ul>} Homo hexameric assembly 5 of PDB entry 4iah coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/4iah_assembly_5_chemically_distinct_molecules_top_image-200x200.png)