Assemblies
Assembly Name:
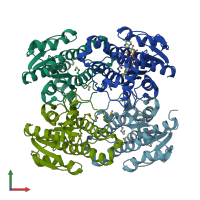
Short-chain dehydrogenase/reductase SDR
Multimeric state:
homo tetramer
Accessible surface area:
31755.58 Å2
Buried surface area:
12451.84 Å2
Dissociation area:
2,659.46
Å2
Dissociation energy (ΔGdiss):
33.97
kcal/mol
Dissociation entropy (TΔSdiss):
14.56
kcal/mol
Symmetry number:
4
PDBe Complex ID:
PDB-CPX-174578
Assembly Name:
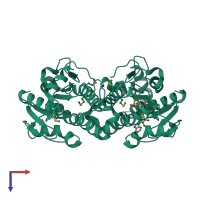
Short-chain dehydrogenase/reductase SDR
Multimeric state:
homo dimer
Accessible surface area:
18327.11 Å2
Buried surface area:
4136.11 Å2
Dissociation area:
1,474.82
Å2
Dissociation energy (ΔGdiss):
21.2
kcal/mol
Dissociation entropy (TΔSdiss):
12.97
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-174577
Assembly Name:
Short-chain dehydrogenase/reductase SDR
Multimeric state:
homo dimer
Accessible surface area:
19109.0 Å2
Buried surface area:
2532.75 Å2
Dissociation area:
1,266.37
Å2
Dissociation energy (ΔGdiss):
10.16
kcal/mol
Dissociation entropy (TΔSdiss):
12.91
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-174577
Assembly Name:
Short-chain dehydrogenase/reductase SDR
Multimeric state:
homo dimer
Accessible surface area:
18747.39 Å2
Buried surface area:
2996.8 Å2
Dissociation area:
1,498.4
Å2
Dissociation energy (ΔGdiss):
22.24
kcal/mol
Dissociation entropy (TΔSdiss):
12.96
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-174577
Assembly Name:
Short-chain dehydrogenase/reductase SDR
Multimeric state:
homo dimer
Accessible surface area:
18851.98 Å2
Buried surface area:
3713.69 Å2
Dissociation area:
1,263.6
Å2
Dissociation energy (ΔGdiss):
10.02
kcal/mol
Dissociation entropy (TΔSdiss):
12.93
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-174577
Macromolecules
Chains: A, B, C, D
Length: 266 amino acids
Theoretical weight: 27.51 KDa
Source organism: Cereibacter sphaeroides 2.4.1
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Enoyl-(Acyl carrier protein) reductase
InterPro:
Length: 266 amino acids
Theoretical weight: 27.51 KDa
Source organism: Cereibacter sphaeroides 2.4.1
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q3IVH6 (Residues: 1-244; Coverage: 100%)
Q3IVH6 (Residues: 1-244; Coverage: 100%)
Pfam: Enoyl-(Acyl carrier protein) reductase
InterPro:
- CGT/MARTX, cysteine protease (CPD) domain superfamily
- NAD(P)-binding domain superfamily
- Short-chain dehydrogenase/reductase SDR