EC 3.6.4.13: RNA helicase
Reaction catalysed:
ATP + H(2)O = ADP + phosphate
Systematic name:
ATP phosphohydrolase (RNA helix unwinding)
Alternative Name(s):
- CSFV NS3 helicase
- DBP2
- DDX17
- DDX25
- DDX3
- DDX3X
- DDX3Y
- DDX4
- DDX5
- DEAD-box RNA helicase
- DEAD-box protein DED1
- DEAH-box RNA helicase
- DEAH-box protein 2
- DED1
- DbpA
- Dex(H/D) RNA helicase
- EIF4A helicase
- EhDEAD1
- EhDEAD1 RNA helicase
- GRTH/DDX25
- KOKV helicase
- Mtr4p
- NPH-II
- Nonstructural protein 3 helicase
- RHA
- RNA helicase A
- RNA helicase DDX3
- RNA helicase Hera
- RNA-dependent ATPase
- TGBp1 NTPase/helicase domain
- VRH1
EC 3.6.1.15: Nucleoside-triphosphate phosphatase
Reaction catalysed:
NTP + H(2)O = NDP + phosphate
Systematic name:
Unspecific diphosphate phosphohydrolase
Alternative Name(s):
- NTPase
- Nucleoside 5-triphosphatase
- Nucleoside triphosphate hydrolase
- Nucleoside triphosphate phosphohydrolase
- Nucleoside-5-triphosphate phosphohydrolase
- Nucleoside-triphosphatase
EC 3.4.21.91: Flavivirin
Reaction catalysed:
Selective hydrolysis of -Xaa-Xaa-|-Yaa- bonds in which each of the Xaa can be either Arg or Lys and Yaa can be either Ser or Ala.
Alternative Name(s):
- NS2B-3 proteinase
- Yellow fever virus (flavivirus) protease
- Yellow fever virus protease
EC 2.7.7.48: RNA-directed RNA polymerase
Reaction catalysed:
Nucleoside triphosphate + RNA(n) = diphosphate + RNA(n+1)
Systematic name:
Nucleoside-triphosphate:RNA nucleotidyltransferase (RNA-directed)
Alternative Name(s):
- 3D polymerase
- PB1 proteins
- PB2 proteins
- Phage f2 replicase
- Polymerase L
- Q-beta replicase
- RDRP
- RNA nucleotidyltransferase (RNA-directed)
- RNA replicase
- RNA synthetase
- RNA transcriptase
- RNA-dependent RNA polymerase
- RNA-dependent RNA replicase
- RNA-dependent ribonucleate nucleotidyltransferase
- Ribonucleic acid replicase
- Ribonucleic acid-dependent ribonucleate nucleotidyltransferase
- Ribonucleic acid-dependent ribonucleic acid polymerase
- Ribonucleic replicase
- Ribonucleic synthetase
- Transcriptase
EC 2.1.1.57: Methyltransferase cap1
Reaction catalysed:
S-adenosyl-L-methionine + a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(ribonucleotide)-[mRNA] = S-adenosyl-L-homocysteine + a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(2'-O-methyl-ribonucleotide)-[mRNA]
Systematic name:
S-adenosyl-L-methionine:5'-(N(7)-methyl 5'-triphosphoguanosine)-(ribonucleotide)-[mRNA] 2-O-methyltransferase
Alternative Name(s):
- Cap1-MTase
- MTR1
- Messenger RNA (nucleoside-2'-)-methyltransferase
- Messenger ribonucleate nucleoside 2'-methyltransferase
- S-adenosyl-L-methionine:mRNA (nucleoside-2'-O)-methyltransferase
- mRNA (nucleoside-2'-O)-methyltransferase
EC 2.1.1.56: mRNA (guanine-N(7))-methyltransferase
Reaction catalysed:
S-adenosyl-L-methionine + a 5'-(5'-triphosphoguanosine)-[mRNA] = S-adenosyl-L-homocysteine + a 5'-(N(7)-methyl 5'-triphosphoguanosine)-[mRNA]
Systematic name:
S-adenosyl-L-methionine:mRNA (guanine-N(7))-methyltransferase
Alternative Name(s):
- Guanine-7-methyltransferase
- Messenger RNA guanine 7-methyltransferase
- Messenger ribonucleate guanine 7-methyltransferase
- S-adenosyl-L-methionine:mRNA (guanine-7-N)-methyltransferase
GO terms
Biochemical function:
Biological process:
- not assigned
Cellular component:
Sequence families
 Protein families (Pfam)
Protein families (Pfam)
 InterPro annotations
InterPro annotations
Structure domains
 CATH domains
CATH domains
4.10.410.10
Class:
Few Secondary Structures
Architecture:
Irregular
Topology:
Factor Xa Inhibitor
Homology:
Pancreatic trypsin inhibitor Kunitz domain
Occurring in:
![The deposited structure of PDB entry <span class='highlight'>3u1j</span> contains 1 copy of CATH domain <span class='highlight'>4.10.410.10 (Factor Xa Inhibitor)</span> in <span class='highlight'>Pancreatic trypsin inhibitor</span>. Showing 1 copy in chain <span class='highlight'>C [auth E]</span>. The deposited structure of PDB entry 3u1j contains 1 copy of CATH domain 4.10.410.10 (Factor Xa Inhibitor) in Pancreatic trypsin inhibitor. Showing 1 copy in chain C [auth E].](https://www.ebi.ac.uk/pdbe/static/entry/3u1j_3_E_CATH_4.10.410.10_image-200x200.png)
2.40.10.10
Class:
Mainly Beta
Architecture:
Beta Barrel
Topology:
Thrombin, subunit H
Homology:
Trypsin-like serine proteases
Occurring in:
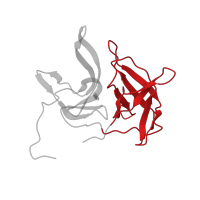



![The deposited structure of PDB entry <span class='highlight'>3u1j</span> contains 1 copy of Pfam domain <span class='highlight'>PF00014 (Kunitz/Bovine pancreatic trypsin inhibitor domain)</span> in <span class='highlight'>Pancreatic trypsin inhibitor</span>. Showing 1 copy in chain <span class='highlight'>C [auth E]</span>. The deposited structure of PDB entry 3u1j contains 1 copy of Pfam domain PF00014 (Kunitz/Bovine pancreatic trypsin inhibitor domain) in Pancreatic trypsin inhibitor. Showing 1 copy in chain C [auth E].](https://www.ebi.ac.uk/pdbe/static/entry/3u1j_3_E_Pfam_PF00014_image-200x200.png)


