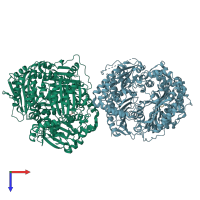
Assemblies
Assembly Name:
Insulin-degrading enzyme and Atrial natriuretic peptide
Multimeric state:
hetero tetramer
Accessible surface area:
76169.19 Å2
Buried surface area:
2978.69 Å2
Dissociation area:
1,387.65
Å2
Dissociation energy (ΔGdiss):
-10.29
kcal/mol
Dissociation entropy (TΔSdiss):
24.96
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-134082
Macromolecules
Chains: A, B
Length: 990 amino acids
Theoretical weight: 114.56 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam:
Length: 990 amino acids
Theoretical weight: 114.56 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P14735 (Residues: 42-1019; Coverage: 96%)
P14735 (Residues: 42-1019; Coverage: 96%)
Pfam:
- Insulinase (Peptidase family M16)
- Peptidase M16 inactive domain
- Middle or third domain of peptidase_M16
- Metalloenzyme, LuxS/M16 peptidase-like
- Peptidase M16, N-terminal
- Peptidase M16, C-terminal
- Peptidase M16, middle/third domain
Chains: C, D
Length: 28 amino acids
Theoretical weight: 3.09 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Atrial natriuretic peptide
InterPro:
Length: 28 amino acids
Theoretical weight: 3.09 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P01160 (Residues: 124-151; Coverage: 22%)
P01160 (Residues: 124-151; Coverage: 22%)
Pfam: Atrial natriuretic peptide
InterPro: