Assemblies
Assembly Name:
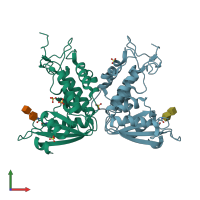
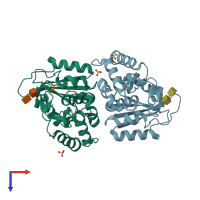
ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1
Multimeric state:
homo dimer
Accessible surface area:
22990.39 Å2
Buried surface area:
3177.5 Å2
Dissociation area:
647.66
Å2
Dissociation energy (ΔGdiss):
4.53
kcal/mol
Dissociation entropy (TΔSdiss):
12.5
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-193579
Assembly Name:
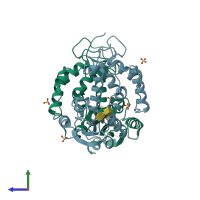
ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1
Multimeric state:
homo dimer
Accessible surface area:
22664.3 Å2
Buried surface area:
3503.59 Å2
Dissociation area:
810.7
Å2
Dissociation energy (ΔGdiss):
-5.5
kcal/mol
Dissociation entropy (TΔSdiss):
12.77
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-193579
Macromolecules
Chains: A, B
Length: 247 amino acids
Theoretical weight: 28.3 KDa
Source organism: Bos taurus
Expression system: Komagataella pastoris
UniProt:
Pfam: ADP-ribosyl cyclase
InterPro: ADP-ribosyl cyclase (CD38/157)
CATH:
Length: 247 amino acids
Theoretical weight: 28.3 KDa
Source organism: Bos taurus
Expression system: Komagataella pastoris
UniProt:
- Canonical:
 Q9TTF5 (Residues: 32-278; Coverage: 98%)
Q9TTF5 (Residues: 32-278; Coverage: 98%)
Pfam: ADP-ribosyl cyclase
InterPro: ADP-ribosyl cyclase (CD38/157)
CATH: