EC 2.7.3.9: Phosphoenolpyruvate--protein phosphotransferase
Reaction catalysed:
Phosphoenolpyruvate + protein L-histidine = pyruvate + protein N(pi)-phospho-L-histidine
Systematic name:
Phosphoenolpyruvate:protein-L-histidine N(pi)-phosphotransferase
Alternative Name(s):
- Enzyme I of the phosphotransferase system
- Phosphoenolpyruvate sugar phosphotransferase enzyme I
- Phosphoenolpyruvate--protein phosphatase
- Phosphoenolpyruvate:protein-L-histidine N-pros-phosphotransferase
- Phosphopyruvate--protein factor phosphotransferase
- Phosphopyruvate--protein phosphotransferase
- Sugar--PEP phosphotransferase enzyme I
GO terms
Biochemical function:
Biological process:
Cellular component:
Sequence families
 Protein families (Pfam)
Protein families (Pfam)
 InterPro annotations
InterPro annotations
IPR002114
Domain description:
Phosphotransferase system, HPr serine phosphorylation site
Occurring in:
Occurring in:
IPR001020
Domain description:
Phosphotransferase system, HPr histidine phosphorylation site
Occurring in:
Occurring in:
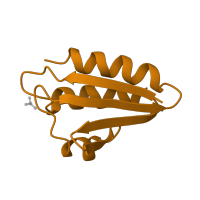
Structure domains
 CATH domains
CATH domains
3.50.30.10
Class:
Alpha Beta
Architecture:
3-Layer(bba) Sandwich
Topology:
Glucose Oxidase; domain 1
Homology:
Phosphohistidine domain
Occurring in:

3.30.1340.10
Class:
Alpha Beta
Architecture:
2-Layer Sandwich
Topology:
Histidine-containing Protein; Chain: A;
Homology:
HPr-like
Occurring in:

 SCOP annotations
SCOP annotations
52013
Class:
Alpha and beta proteins (a/b)
Fold:
The "swivelling" beta/beta/alpha domain
Superfamily:
Phosphohistidine domain
Occurring in:

47832
Class:
All alpha proteins
Fold:
SAM domain-like
Superfamily:
Enzyme I of the PEP:sugar phosphotransferase system HPr-binding (sub)domain
Occurring in:







