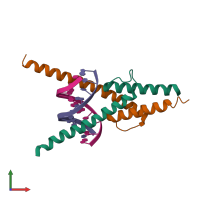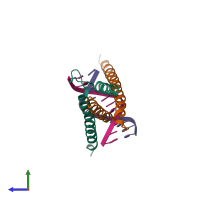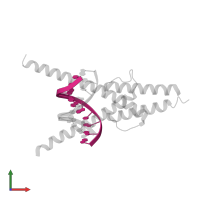Assemblies
Assembly Name:
Transcription factor E2-alpha and T-cell acute lymphocytic leukemia protein 1 and DNA
Multimeric state:
hetero tetramer
Accessible surface area:
12936.28 Å2
Buried surface area:
5511.74 Å2
Dissociation area:
1,213.58
Å2
Dissociation energy (ΔGdiss):
18.31
kcal/mol
Dissociation entropy (TΔSdiss):
10.96
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-115952
Macromolecules
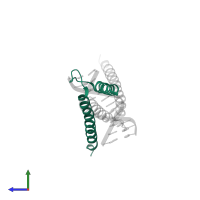
Chain: A
Length: 91 amino acids
Theoretical weight: 10.72 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Helix-loop-helix DNA-binding domain
InterPro:
Length: 91 amino acids
Theoretical weight: 10.72 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P17542 (Residues: 180-253; Coverage: 22%)
P17542 (Residues: 180-253; Coverage: 22%)
Pfam: Helix-loop-helix DNA-binding domain
InterPro:
- Helix-loop-helix protein TAL-like
- Helix-loop-helix DNA-binding domain superfamily
- Myc-type, basic helix-loop-helix (bHLH) domain
Chain: B
Length: 82 amino acids
Theoretical weight: 9.67 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Helix-loop-helix DNA-binding domain
InterPro:
CATH: Helix-loop-helix DNA-binding domain
Length: 82 amino acids
Theoretical weight: 9.67 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P15923 (Residues: 538-616; Coverage: 12%)
P15923 (Residues: 538-616; Coverage: 12%)
Pfam: Helix-loop-helix DNA-binding domain
InterPro:
CATH: Helix-loop-helix DNA-binding domain