Assemblies
Assembly Name:
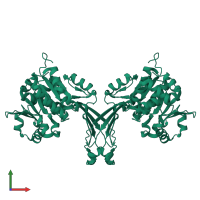
Adenosine kinase
Multimeric state:
homo dimer
Accessible surface area:
27582.23 Å2
Buried surface area:
3120.73 Å2
Dissociation area:
1,560.37
Å2
Dissociation energy (ΔGdiss):
11.68
kcal/mol
Dissociation entropy (TΔSdiss):
13.81
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-161618
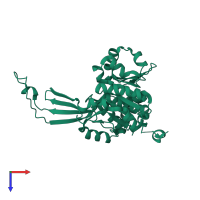
Assembly Name:
Adenosine kinase
Multimeric state:
monomeric
Accessible surface area:
15880.63 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-161617
Assembly Name:
Adenosine kinase
Multimeric state:
homo dimer
Accessible surface area:
28833.28 Å2
Buried surface area:
2940.62 Å2
Dissociation area:
1,470.31
Å2
Dissociation energy (ΔGdiss):
10.54
kcal/mol
Dissociation entropy (TΔSdiss):
13.83
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-161618
Macromolecules
Chains: A, B
Length: 334 amino acids
Theoretical weight: 35.68 KDa
Source organism: Mycobacterium tuberculosis
Expression system: Escherichia coli
UniProt:
Pfam: pfkB family carbohydrate kinase
InterPro:
CATH: UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase
Length: 334 amino acids
Theoretical weight: 35.68 KDa
Source organism: Mycobacterium tuberculosis
Expression system: Escherichia coli
UniProt:
- Canonical:
 P9WID5 (Residues: 1-324; Coverage: 100%)
P9WID5 (Residues: 1-324; Coverage: 100%)
Pfam: pfkB family carbohydrate kinase
InterPro:
CATH: UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase