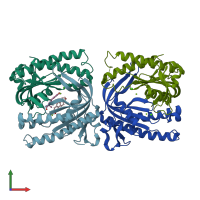
Assemblies
Assembly Name:
histidine kinase
Multimeric state:
homo dimer
Accessible surface area:
16664.6 Å2
Buried surface area:
6340.43 Å2
Dissociation area:
1,981.4
Å2
Dissociation energy (ΔGdiss):
8.81
kcal/mol
Dissociation entropy (TΔSdiss):
13.01
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-128446
Assembly Name:
histidine kinase
Multimeric state:
homo dimer
Accessible surface area:
16459.67 Å2
Buried surface area:
5435.09 Å2
Dissociation area:
1,988.48
Å2
Dissociation energy (ΔGdiss):
6.78
kcal/mol
Dissociation entropy (TΔSdiss):
12.98
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-128446
Macromolecules
Chains: A, B, C, D
Length: 219 amino acids
Theoretical weight: 24.19 KDa
Source organism: Arthrospira platensis
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Adenylate and Guanylate cyclase catalytic domain
InterPro:
Length: 219 amino acids
Theoretical weight: 24.19 KDa
Source organism: Arthrospira platensis
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 O32393 (Residues: 1004-1202; Coverage: 17%)
O32393 (Residues: 1004-1202; Coverage: 17%)
Pfam: Adenylate and Guanylate cyclase catalytic domain
InterPro:
- 6-pyruvoyl tetrahydropterin synthase/QueD superfamily
- Adenylyl cyclase class-3/4/guanylyl cyclase
- Nucleotide cyclase