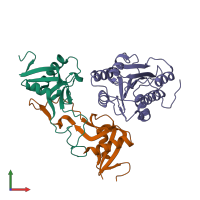
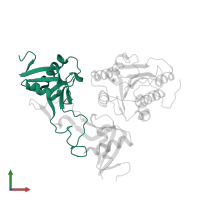
Assemblies
Multimeric state:
hetero trimer
Accessible surface area:
20134.62 Å2
Buried surface area:
5032.24 Å2
Dissociation area:
874.6
Å2
Dissociation energy (ΔGdiss):
-3.51
kcal/mol
Dissociation entropy (TΔSdiss):
12.52
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-137569
Macromolecules
Chain: A
Length: 131 amino acids
Theoretical weight: 14.95 KDa
Source organism: Bitis arietans
UniProt:
InterPro:
CATH: Mannose-Binding Protein A, subunit A
SCOP: C-type lectin domain
Length: 131 amino acids
Theoretical weight: 14.95 KDa
Source organism: Bitis arietans
UniProt:
- Canonical:
 Q7LZK5 (Residues: 1-131; Coverage: 100%)
Q7LZK5 (Residues: 1-131; Coverage: 100%)
InterPro:
CATH: Mannose-Binding Protein A, subunit A
SCOP: C-type lectin domain
Chain: B
Length: 125 amino acids
Theoretical weight: 14.82 KDa
Source organism: Bitis arietans
UniProt:
InterPro:
CATH: Mannose-Binding Protein A, subunit A
SCOP: C-type lectin domain
Length: 125 amino acids
Theoretical weight: 14.82 KDa
Source organism: Bitis arietans
UniProt:
- Canonical:
 Q7LZK8 (Residues: 1-125; Coverage: 100%)
Q7LZK8 (Residues: 1-125; Coverage: 100%)
InterPro:
CATH: Mannose-Binding Protein A, subunit A
SCOP: C-type lectin domain
Chain: C
Length: 209 amino acids
Theoretical weight: 23.89 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: von Willebrand factor type A domain
InterPro:
CATH: von Willebrand factor, type A domain
SCOP: Integrin A (or I) domain
Length: 209 amino acids
Theoretical weight: 23.89 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P04275 (Residues: 1260-1468; Coverage: 8%)
P04275 (Residues: 1260-1468; Coverage: 8%)
Pfam: von Willebrand factor type A domain
InterPro:
CATH: von Willebrand factor, type A domain
SCOP: Integrin A (or I) domain