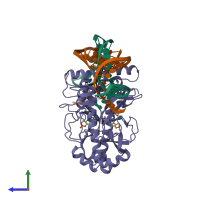
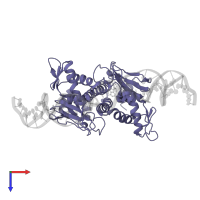
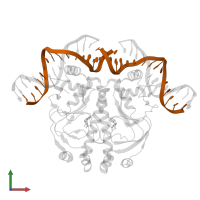
Assemblies
Assembly Name:
DNA-binding transcriptional dual regulator CRP and DNA
Multimeric state:
hetero hexamer
Accessible surface area:
27248.38 Å2
Buried surface area:
10943.48 Å2
Dissociation area:
1,163.41
Å2
Dissociation energy (ΔGdiss):
12.84
kcal/mol
Dissociation entropy (TΔSdiss):
12.41
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-114543
Macromolecules
Chains: A, B
Length: 209 amino acids
Theoretical weight: 23.56 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
SCOP:
Length: 209 amino acids
Theoretical weight: 23.56 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
- Canonical:
 P0ACJ8 (Residues: 2-210; Coverage: 100%)
P0ACJ8 (Residues: 2-210; Coverage: 100%)
Pfam:
InterPro:
- Cyclic nucleotide-binding domain
- RmlC-like jelly roll fold
- Cyclic nucleotide-binding domain superfamily
- Cyclic nucleotide-binding, conserved site
- Crp-type HTH domain
- Winged helix-like DNA-binding domain superfamily
- Winged helix DNA-binding domain superfamily
- Transcription regulator HTH, Crp-type, conserved site
SCOP:
Name:
DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3')
Representative chains: C, E
Source organism: Escherichia coli [562]
Expression system: Not provided
Length: 14 nucleotides
Theoretical weight: 4.35 KDa
Representative chains: C, E
Source organism: Escherichia coli [562]
Expression system: Not provided
Length: 14 nucleotides
Theoretical weight: 4.35 KDa
Name:
DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3')
Representative chains: D, F
Source organism: Escherichia coli [562]
Expression system: Not provided
Length: 17 nucleotides
Theoretical weight: 5.15 KDa
Representative chains: D, F
Source organism: Escherichia coli [562]
Expression system: Not provided
Length: 17 nucleotides
Theoretical weight: 5.15 KDa