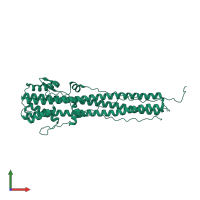
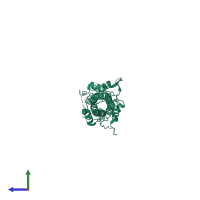
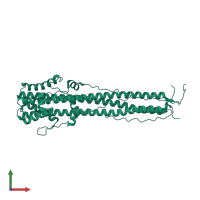
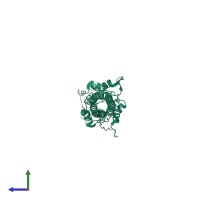
Assemblies
Assembly Name:
Hemagglutinin HA1 chain
Multimeric state:
homo trimer
Accessible surface area:
21765.23 Å2
Buried surface area:
12619.31 Å2
Dissociation area:
6,309.65
Å2
Dissociation energy (ΔGdiss):
91.17
kcal/mol
Dissociation entropy (TΔSdiss):
27.53
kcal/mol
Symmetry number:
3
PDBe Complex ID:
PDB-CPX-137029
Assembly Name:
Hemagglutinin HA1 chain
Multimeric state:
homo trimer
Accessible surface area:
22684.53 Å2
Buried surface area:
13178.21 Å2
Dissociation area:
6,589.11
Å2
Dissociation energy (ΔGdiss):
94.86
kcal/mol
Dissociation entropy (TΔSdiss):
27.78
kcal/mol
Symmetry number:
3
PDBe Complex ID:
PDB-CPX-137029
Macromolecules
Chains: A, B, C, D, E, F
Length: 155 amino acids
Theoretical weight: 18.06 KDa
Source organism: Influenza A virus
Expression system: Escherichia coli
UniProt:
InterPro:
CATH: Hemagglutinin Ectodomain; Chain B
SCOP: Influenza hemagglutinin (stalk)
Length: 155 amino acids
Theoretical weight: 18.06 KDa
Source organism: Influenza A virus
Expression system: Escherichia coli
UniProt:
- Canonical:
 P03437 (Residues: 376-530; Coverage: 28%)
P03437 (Residues: 376-530; Coverage: 28%)
InterPro:
CATH: Hemagglutinin Ectodomain; Chain B
SCOP: Influenza hemagglutinin (stalk)