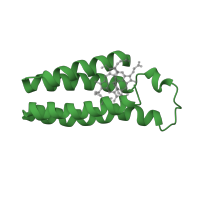
THE SOLUTION STRUCTURE OF THE HEME BINDING VARIANT ARG98CYS OF OXIDIZED ESCHERICHIA COLI CYTOCHROME B562
Source organism:
Escherichia coli
Biochemical function:
metal ion binding
Biological process: electron transport chain
Cellular component: periplasmic space
Biological process: electron transport chain
Cellular component: periplasmic space