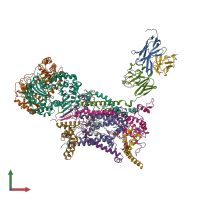
Function and Biology Details
Reaction catalysed:
Quinol + 2 ferricytochrome c(Side 2) = quinone + 2 ferrocytochrome c(Side 2) + 2 H(+)(Side 2)
Biochemical function:
Biological process:
Cellular component:
Sequence domains:
Structure domains:
- MPP-like
- V set domains (antibody variable domain-like)
- Subunit X (non-heme 7 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
- Ubiquinone-binding protein QP-C of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
- 14 kDa protein of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
- Non-heme 11 kDa protein of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
- Rieske iron-sulfur protein (ISP)
- ISP transmembrane anchor
4 more domains
Structure analysis Details
Assemblies composition:
PDBe Complex ID:
PDB-CPX-132277 (preferred)
Entry contents:
11 distinct polypeptide molecules
Macromolecules (11 distinct):